Mechanism of BRCA1-BARD1 function in DNA end resection and DNA protection
- PMID: 39261728
- PMCID: PMC11464378
- DOI: 10.1038/s41586-024-07909-9
Mechanism of BRCA1-BARD1 function in DNA end resection and DNA protection
Abstract
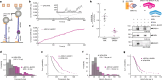
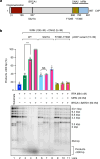
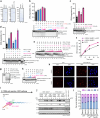
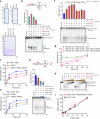
DNA double-strand break (DSB) repair by homologous recombination is initiated by DNA end resection, a process involving the controlled degradation of the 5'-terminated strands at DSB sites1,2. The breast cancer suppressor BRCA1-BARD1 not only promotes resection and homologous recombination, but it also protects DNA upon replication stress1,3-9. BRCA1-BARD1 counteracts the anti-resection and pro-non-homologous end-joining factor 53BP1, but whether it functions in resection directly has been unclear10-16. Using purified recombinant proteins, we show here that BRCA1-BARD1 directly promotes long-range DNA end resection pathways catalysed by the EXO1 or DNA2 nucleases. In the DNA2-dependent pathway, BRCA1-BARD1 stimulates DNA unwinding by the Werner or Bloom helicase. Together with MRE11-RAD50-NBS1 and phosphorylated CtIP, BRCA1-BARD1 forms the BRCA1-C complex17,18, which stimulates resection synergistically to an even greater extent. A mutation in phosphorylated CtIP (S327A), which disrupts its binding to the BRCT repeats of BRCA1 and hence the integrity of the BRCA1-C complex19-21, inhibits resection, showing that BRCA1-C is a functionally integrated ensemble. Whereas BRCA1-BARD1 stimulates resection in DSB repair, it paradoxically also protects replication forks from unscheduled degradation upon stress, which involves a homologous recombination-independent function of the recombinase RAD51 (refs. 4-6,8). We show that in the presence of RAD51, BRCA1-BARD1 instead inhibits DNA degradation. On the basis of our data, the presence and local concentration of RAD51 might determine the balance between the pronuclease and the DNA protection functions of BRCA1-BARD1 in various physiological contexts.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures












Similar articles
-
Promotion of DNA end resection by BRCA1-BARD1 in homologous recombination.Nature. 2024 Oct;634(8033):482-491. doi: 10.1038/s41586-024-07910-2. Epub 2024 Sep 11. Nature. 2024. PMID: 39261729
-
Super-resolution mapping of cellular double-strand break resection complexes during homologous recombination.Proc Natl Acad Sci U S A. 2021 Mar 16;118(11):e2021963118. doi: 10.1073/pnas.2021963118. Proc Natl Acad Sci U S A. 2021. PMID: 33707212 Free PMC article.
-
CtIP promotes the motor activity of DNA2 to accelerate long-range DNA end resection.Proc Natl Acad Sci U S A. 2020 Apr 21;117(16):8859-8869. doi: 10.1073/pnas.2001165117. Epub 2020 Apr 2. Proc Natl Acad Sci U S A. 2020. PMID: 32241893 Free PMC article.
-
The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication.Nat Rev Mol Cell Biol. 2020 May;21(5):284-299. doi: 10.1038/s41580-020-0218-z. Epub 2020 Feb 24. Nat Rev Mol Cell Biol. 2020. PMID: 32094664 Free PMC article. Review.
-
Mechanism and regulation of DNA end resection in eukaryotes.Crit Rev Biochem Mol Biol. 2016 May-Jun;51(3):195-212. doi: 10.3109/10409238.2016.1172552. Epub 2016 Apr 20. Crit Rev Biochem Mol Biol. 2016. PMID: 27098756 Free PMC article. Review.
Cited by
-
Promotion of DNA end resection by BRCA1-BARD1 in homologous recombination.Nature. 2024 Oct;634(8033):482-491. doi: 10.1038/s41586-024-07910-2. Epub 2024 Sep 11. Nature. 2024. PMID: 39261729
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

