Drug repurposing for Parkinson's disease by biological pathway based edge-weighted network proximity analysis
- PMID: 39261542
- PMCID: PMC11390918
- DOI: 10.1038/s41598-024-71922-1
Drug repurposing for Parkinson's disease by biological pathway based edge-weighted network proximity analysis
Abstract
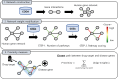
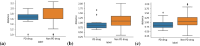
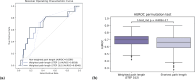
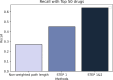
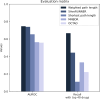
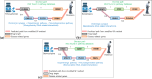
Parkinson's disease is the second most frequent neurodegenerative disease, and its severity is increasing with extended life expectancy. Most of current treatments provide symptomatic relief; however, disease progression is not inhibited. There are multiple trials for treatments that target the causes of the disease but they were flawed. The mechanisms underlying neurodegenerative diseases are intricate, and understanding the interplay among the biological elements involved is crucial. These relationships can be effectively analyzed through biological networks, and the application of network-based analyses in the context of neurodegenerative disease treatment has gained considerable attention. Moreover, considering the significance differences in interactions between biological elements within the network is important. Therefore, we introduce a novel biological pathway based edge-weighted network construction method for drug repurposing in Parkinson's disease. The interaction found in multiple Parkinson's disease-related pathways is more significant than other interactions, and this significance is reflected in the network edge weights. Using the edge-weighted network construction method, we found a significant difference in the efficacy between known and unknown Parkinson's disease drugs. The method predicts drug-disease interactions more accurately than approaches that do not consider the significance differences among interactions, and the paths between the drug and disease within the network correspond to the drug's mechanism of action. In summary, we propose a network-based drug repurposing method using the biological pathway based edge-weighted network. Using this methodology, researchers can find novel drug candidates for the parkinson's disease and their mechanism of actions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
A bidirectional drug repositioning approach for Parkinson's disease through network-based inference.Biochem Biophys Res Commun. 2015 Feb 13;457(3):280-7. doi: 10.1016/j.bbrc.2014.12.101. Epub 2015 Jan 7. Biochem Biophys Res Commun. 2015. PMID: 25576361
-
Drug Repurposing in Parkinson's Disease.CNS Drugs. 2018 Aug;32(8):747-761. doi: 10.1007/s40263-018-0548-y. CNS Drugs. 2018. PMID: 30066310 Review.
-
Advances in Parkinson's disease research - A computational network pharmacological approach.Int Immunopharmacol. 2024 Sep 30;139:112758. doi: 10.1016/j.intimp.2024.112758. Epub 2024 Jul 26. Int Immunopharmacol. 2024. PMID: 39067399 Review.
-
Recent Advances in Drug Repurposing for Parkinson's Disease.Curr Med Chem. 2019;26(28):5340-5362. doi: 10.2174/0929867325666180719144850. Curr Med Chem. 2019. PMID: 30027839 Review.
-
Tetracycline repurposing in neurodegeneration: focus on Parkinson's disease.J Neural Transm (Vienna). 2018 Oct;125(10):1403-1415. doi: 10.1007/s00702-018-1913-1. Epub 2018 Aug 14. J Neural Transm (Vienna). 2018. PMID: 30109452 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

