This is a preprint.
Symbiotic bacteria Sodalis glossinidius, Spiroplasma sp and Wolbachia do not favour Trypanosoma grayi coexistence in wild population of tsetse flies collected in Bobo-Dioulasso, Burkina Faso
- PMID: 39257987
- PMCID: PMC11384793
- DOI: 10.21203/rs.3.rs-4756528/v1
Symbiotic bacteria Sodalis glossinidius, Spiroplasma sp and Wolbachia do not favour Trypanosoma grayi coexistence in wild population of tsetse flies collected in Bobo-Dioulasso, Burkina Faso
Update in
-
Symbiotic bacteria Sodalis glossinidius, Spiroplasma sp and Wolbachia do not favour Trypanosoma grayi coexistence in wild population of tsetse flies collected in Bobo-Dioulasso, Burkina Faso.BMC Microbiol. 2024 Sep 28;24(1):373. doi: 10.1186/s12866-024-03531-x. BMC Microbiol. 2024. PMID: 39342132 Free PMC article.
Abstract
Background: Tsetse flies, the biological vectors of African trypanosomes, have established symbiotic associations with different bacteria. Their vector competence is suggested to be affected by bacterial endosymbionts. The current study provided the prevalence of three tsetse symbiotic bacteria and trypanosomes in Glossina species from Burkina Faso.
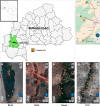
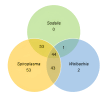
Results: A total of 430 tsetse flies were captured using biconical traps in four different collection sites around Bobo-Dioulasso (Bama, Bana, Nasso, and Peni), and their guts were removed. Two hundred tsetse were randomly selected and their guts were screened byPCR for the presence of Sodalis glossinidius, Spiroplasmasp., Wolbachia and trypanosomes. Of the 200 tsetse, 196 (98.0%) were Glossina palpalis gambienseand 4 (2.0%) Glossina tachinoides. The overall symbiont prevalence was 49.0%, 96.5%, and 45.0%, respectively for S. glossinidius, Spiroplasma and Wolbachia. Prevalence varied between sampling locations: S. glossinidius(54.7%, 38.5%, 31.6%, 70.8%); Spiroplasma (100%, 100%, 87.7%, 100%); and Wolbachia(43.4%, 38.5%, 38.6%, 70.8%),respectively in Bama, Bana, Nasso and Peni. Noteworthy, no G. tachhnoideswas infected by S. glossinidius and Wolbachia, but they were all infected by Spiroplasma sp. A total of 196 (98.0 %) harbored at least one endosymbionts. Fifty-five (27.5%) carried single endosymbiont. Trypanosomes were found only in G.p. gambiense, but not G. tachinoides. Trypanosomes were present in flies from all study locations with an overall prevalence of 29.5%. In Bama, Bana, Nasso, and Peni, the trypanosome infection rate was respectively 39.6%, 23.1%, 8.8%, and 37.5%. Remarkably, only Trypanosoma grayi was present. Of all trypanosome-infected flies, 55.9%, 98.3%, and 33.9% hosted S. glossinidius, Spiroplasma sp and Wolbachia, respectively. There was no association between Sodalis, Spiroplasma and trypanosome presence, but there was a negative association with Wolbachia presence. We reported1.9 times likelihood of trypanosome absence when Wolbachia was present.
Conclusion: This is the first survey reporting the presence of Trypanosoma grayi in tsetse from Burkina Faso. Tsetse from these localities were highly positive for symbiotic bacteria, more predominantly with Spiroplasma sp. Modifications of symbiotic interactions may pave way for disease control.
Keywords: Burkina Faso; Sodalis glossinidius; Spiroplasma; Trypanosoma grayi; Tsetse flies; Wolbachia.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures




Similar articles
-
Symbiotic bacteria Sodalis glossinidius, Spiroplasma sp and Wolbachia do not favour Trypanosoma grayi coexistence in wild population of tsetse flies collected in Bobo-Dioulasso, Burkina Faso.BMC Microbiol. 2024 Sep 28;24(1):373. doi: 10.1186/s12866-024-03531-x. BMC Microbiol. 2024. PMID: 39342132 Free PMC article.
-
Molecular detection of Sodalis glossinidius, Spiroplasma species and Wolbachia endosymbionts in wild population of tsetse flies collected in Cameroon, Chad and Nigeria.BMC Microbiol. 2023 Sep 16;23(1):260. doi: 10.1186/s12866-023-03005-6. BMC Microbiol. 2023. PMID: 37716961 Free PMC article.
-
Prevalence of trypanosomes, salivary gland hypertrophy virus and Wolbachia in wild populations of tsetse flies from West Africa.BMC Microbiol. 2018 Nov 23;18(Suppl 1):153. doi: 10.1186/s12866-018-1287-4. BMC Microbiol. 2018. PMID: 30470187 Free PMC article.
-
Tsetse-Wolbachia symbiosis: comes of age and has great potential for pest and disease control.J Invertebr Pathol. 2013 Mar;112 Suppl(0):S94-103. doi: 10.1016/j.jip.2012.05.010. Epub 2012 Jul 23. J Invertebr Pathol. 2013. PMID: 22835476 Free PMC article. Review.
-
Bacterial Symbionts of Tsetse Flies: Relationships and Functional Interactions Between Tsetse Flies and Their Symbionts.Results Probl Cell Differ. 2020;69:497-536. doi: 10.1007/978-3-030-51849-3_19. Results Probl Cell Differ. 2020. PMID: 33263885 Review.
References
-
- Swallow BM. Impacts of Trypanosomiasis on African agriculture. PAAT technical and scientific series 2. 2000.
-
- Lindh JM, Lehane MJ. The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek. 2011;99(3):711–20. - PubMed
-
- Mfopit YM, Achukwi DM, Mamman M, Balogun EO, Shuaibu MN, Kabir J. Isolation, Characterization and Antimicrobial Susceptibility Pattern of Pseudomonas aeruginosa from Tsetse Flies Captured in Yankari Game Reserve, Nigeria. Eur J Med Health Sci. 2023;5(4):94–9.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

