Soil microbiome characterization and its future directions with biosensing
- PMID: 39256848
- PMCID: PMC11389470
- DOI: 10.1186/s13036-024-00444-1
Soil microbiome characterization and its future directions with biosensing
Abstract
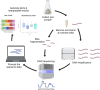
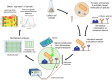
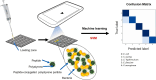
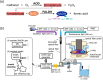
Soil microbiome characterization is typically achieved with next-generation sequencing (NGS) techniques. Metabarcoding is very common, and meta-omics is growing in popularity. These techniques have been instrumental in microbiology, but they have limitations. They require extensive time, funding, expertise, and computing power to be effective. Moreover, these techniques are restricted to controlled laboratory conditions; they are not applicable in field settings, nor can they rapidly generate data. This hinders using NGS as an environmental monitoring tool or an in-situ checking device. Biosensing technology can be applied to soil microbiome characterization to overcome these limitations and to complement NGS techniques. Biosensing has been used in biomedical applications for decades, and many successful commercial products are on the market. Given its previous success, biosensing has much to offer soil microbiome characterization. There is a great variety of biosensors and biosensing techniques, and a few in particular are better suited for soil field studies. Aptamers are more stable than enzymes or antibodies and are more ready for field-use biosensors. Given that any microbiome is complex, a multiplex sensor will be needed, and with large, complicated datasets, machine learning might benefit these analyses. If the signals from the biosensors are optical, a smartphone can be used as a portable optical reader and potential data-analyzing device. Biosensing is a rich field that couples engineering and biology, and applying its toolset to help advance soil microbiome characterization would be a boon to microbiology more broadly.
Keywords: Aptamer; Biosensor; Machine learning; Soil health; Soil microbiome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Aptamer-based biosensors for biomedical diagnostics.Analyst. 2014 Jun 7;139(11):2627-40. doi: 10.1039/c4an00132j. Analyst. 2014. PMID: 24733714 Review.
-
The Uncharted Microbial World: Microbes and Their Activities in the Environment: This report is based on a colloquium, sponsored by the American Academy of Microbiology, convened February 9-11, 2007, in Seattle, Washington.Washington (DC): American Society for Microbiology; 2008. Washington (DC): American Society for Microbiology; 2008. PMID: 33001603 Free Books & Documents. Review.
-
Nanostructures for Biosensing, with a Brief Overview on Cancer Detection, IoT, and the Role of Machine Learning in Smart Biosensors.Sensors (Basel). 2021 Feb 10;21(4):1253. doi: 10.3390/s21041253. Sensors (Basel). 2021. PMID: 33578726 Free PMC article. Review.
-
Recent advances in soil microbial fuel cells based self-powered biosensor.Chemosphere. 2022 Sep;303(Pt 1):135036. doi: 10.1016/j.chemosphere.2022.135036. Epub 2022 May 21. Chemosphere. 2022. PMID: 35609665 Review.
-
Erratum: Eyestalk Ablation to Increase Ovarian Maturation in Mud Crabs.J Vis Exp. 2023 May 26;(195). doi: 10.3791/6561. J Vis Exp. 2023. PMID: 37235796
References
-
- Coyne MS. A cartoon history of soil microbiology. J Nat Res Life Sci Educ. 1996;25:30–6. 10.2134/jnrlse.1996.0030.10.2134/jnrlse.1996.0030 - DOI
-
- Durán P, Jorquera M, Viscardi S, Carrion VJ, Mora M de la L, Pozo MJ. Screening and characterization of potentially suppressive soils against Gaeumannomyces graminis under extensive wheat cropping by Chilean indigenous communities. Front Microbiol. 2017;8:1552. 10.3389/fmicb.2017.01552. - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

