FABP4 deficiency ameliorates alcoholic steatohepatitis in mice via inhibition of p53 signaling pathway
- PMID: 39256510
- PMCID: PMC11387727
- DOI: 10.1038/s41598-024-71311-8
FABP4 deficiency ameliorates alcoholic steatohepatitis in mice via inhibition of p53 signaling pathway
Abstract
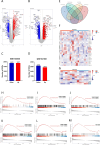
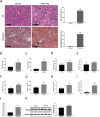
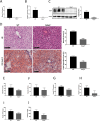
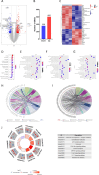
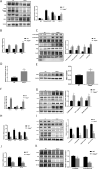
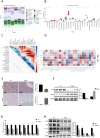
Fatty acid-binding protein 4 (FABP4) plays an essential role in metabolism and inflammation. However, the role of FABP4 in alcoholic steatohepatitis (ASH) remains unclear. This study aimed to investigate the function and underlying mechanisms of FABP4 in the progression of ASH. We first obtained alcoholic hepatitis (AH) datasets from the National Center for Biotechnology Information-Gene Expression Omnibus database and conducted bioinformatics analysis to identify critical genes in the FABP family. We then established ASH models of the wild-type (WT) and Fabp4-deficient (Fabp4-/-) mice to investigate the role of FABP4 in ASH. Additionally, we performed transcriptional profiling of mouse liver tissue and analyzed the results using integrative bioinformatics. The FABP4-associated signaling pathway was further verified. FABP4 was upregulated in two AH datasets and was thus identified as a critical biomarker for AH. FABP4 expression was higher in the liver tissues of patients with alcoholic liver disease and ASH mice than in the corresponding control samples. Furthermore, the Fabp4-/- ASH mice showed reduced hepatic lipid deposition and inflammation compared with the WT ASH mice. Mechanistically, Fabp4 may be involved in regulating the p53 and sirtuin-1 signaling pathways, subsequently affecting lipid metabolism and macrophage polarization in the liver of ASH mice. Our results demonstrate that Fabp4 is involved in the progression of ASH and that Fabp4 deficiency may ameliorate ASH. Therefore, FABP4 may be a potential therapeutic target for ASH treatment.
Keywords: Alcoholic steatohepatitis (ASH); Bioinformatics analysis; FABP4; p53.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Fat-Specific Protein 27/CIDEC Promotes Development of Alcoholic Steatohepatitis in Mice and Humans.Gastroenterology. 2015 Oct;149(4):1030-41.e6. doi: 10.1053/j.gastro.2015.06.009. Epub 2015 Jun 20. Gastroenterology. 2015. PMID: 26099526 Free PMC article.
-
PARP inhibition protects against alcoholic and non-alcoholic steatohepatitis.J Hepatol. 2017 Mar;66(3):589-600. doi: 10.1016/j.jhep.2016.10.023. Epub 2016 Oct 29. J Hepatol. 2017. PMID: 27984176
-
Deletion of SIRT1 from hepatocytes in mice disrupts lipin-1 signaling and aggravates alcoholic fatty liver.Gastroenterology. 2014 Mar;146(3):801-11. doi: 10.1053/j.gastro.2013.11.008. Epub 2013 Nov 18. Gastroenterology. 2014. PMID: 24262277 Free PMC article.
-
Signal Transduction Mechanisms of Alcoholic Fatty Liver Disease: Emer ging Role of Lipin-1.Curr Mol Pharmacol. 2017;10(3):226-236. doi: 10.2174/1874467208666150817112109. Curr Mol Pharmacol. 2017. PMID: 26278388 Free PMC article. Review.
-
p53 as a double-edged sword in the progression of non-alcoholic fatty liver disease.Life Sci. 2018 Dec 15;215:64-72. doi: 10.1016/j.lfs.2018.10.051. Epub 2018 Oct 26. Life Sci. 2018. PMID: 30473026 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

