The hidden base of the iceberg: gut peptidoglycome dynamics is foundational to its influence on the host
- PMID: 39239828
- PMCID: PMC11382707
- DOI: 10.1080/19490976.2024.2395099
The hidden base of the iceberg: gut peptidoglycome dynamics is foundational to its influence on the host
Abstract
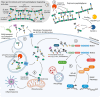
The intestinal microbiota of humans includes a highly diverse range of bacterial species. All these bacteria possess a cell wall, composed primarily of the macromolecule peptidoglycan. As such, the gut also harbors an abundant and varied peptidoglycome. A remarkable range of host physiological pathways are regulated by peptidoglycan fragments that originate from the gut microbiota and enter the host system. Interactions between the host system and peptidoglycan can influence physiological development and homeostasis, promote health, or contribute to inflammatory disease. Underlying these effects is the interplay between microbiota composition and enzymatic processes that shape the intestinal peptidoglycome, dictating the types of peptidoglycan generated, that subsequently cross the gut barrier. In this review, we highlight and discuss the hidden and emerging functional aspects of the microbiome, i.e. the hidden base of the iceberg, that modulate the composition of gut peptidoglycan, and how these fundamental processes are drivers of physiological outcomes for the host.
Keywords: Gut; Microbiota; PG trafficking; chronic inflammation; host signaling; peptidoglycan; peptidoglycan processing; prophage.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



Similar articles
-
Microbiota-induced active translocation of peptidoglycan across the intestinal barrier dictates its within-host dissemination.Proc Natl Acad Sci U S A. 2023 Jan 24;120(4):e2209936120. doi: 10.1073/pnas.2209936120. Epub 2023 Jan 20. Proc Natl Acad Sci U S A. 2023. PMID: 36669110 Free PMC article.
-
Uptake, recognition and responses to peptidoglycan in the mammalian host.FEMS Microbiol Rev. 2021 Jan 8;45(1):fuaa044. doi: 10.1093/femsre/fuaa044. FEMS Microbiol Rev. 2021. PMID: 32897324 Free PMC article. Review.
-
Impact of Bacterial Metabolites on Gut Barrier Function and Host Immunity: A Focus on Bacterial Metabolism and Its Relevance for Intestinal Inflammation.Front Immunol. 2021 May 26;12:658354. doi: 10.3389/fimmu.2021.658354. eCollection 2021. Front Immunol. 2021. PMID: 34122415 Free PMC article. Review.
-
Microbiota-derived extracellular vesicles in interkingdom communication in the gut.J Extracell Vesicles. 2021 Nov;10(13):e12161. doi: 10.1002/jev2.12161. J Extracell Vesicles. 2021. PMID: 34738337 Free PMC article. Review.
-
Host-microbiota interaction in intestinal stem cell homeostasis.Gut Microbes. 2024 Jan-Dec;16(1):2353399. doi: 10.1080/19490976.2024.2353399. Epub 2024 May 17. Gut Microbes. 2024. PMID: 38757687 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
