This is a preprint.
Identifying novel chemical matter against the Chikungunya virus nsP3 macrodomain through crystallographic fragment screening
- PMID: 39229067
- PMCID: PMC11370605
- DOI: 10.1101/2024.08.23.609196
Identifying novel chemical matter against the Chikungunya virus nsP3 macrodomain through crystallographic fragment screening
Abstract
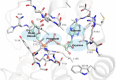
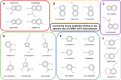
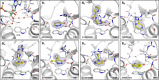
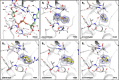
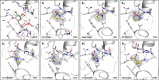
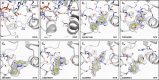
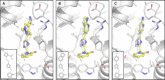
Chikungunya virus (CHIKV) causes severe fever, rash and debilitating joint pain that can last for months1,2or even years. Millions of people have been infected with CHIKV, mostly in low and middle-income countries, and the virus continues to spread into new areas due to the geographical expansion of its mosquito hosts. Its genome encodes a macrodomain, which functions as an ADP-ribosyl hydrolase, removing ADPr from viral and host-cell proteins interfering with the innate immune response. Mutational studies have shown that the CHIKV nsP3 macrodomain is necessary for viral replication, making it a potential target for the development of antiviral therapeutics. We, therefore, performed a high-throughput crystallographic fragment screen against the CHIKV nsP3 macrodomain, yielding 109 fragment hits covering the ADPr-binding site and two adjacent subsites that are absent in the homologous macrodomain of SARS-CoV-2 but may be present in other alphaviruses, such as Venezuelan equine encephalitis virus (VEEV) and eastern equine encephalitis virus (EEEV). Finally, a subset of overlapping fragments was used to manually design three fragment merges covering the adenine and oxyanion subsites. The rich dataset of chemical matter and structural information discovered from this fragment screen is publicly available and can be used as a starting point for developing a CHIKV nsP3 macrodomain inhibitor.
Figures


Similar articles
-
ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication.Proc Natl Acad Sci U S A. 2018 Oct 30;115(44):E10457-E10466. doi: 10.1073/pnas.1812130115. Epub 2018 Oct 15. Proc Natl Acad Sci U S A. 2018. PMID: 30322911 Free PMC article.
-
Both ADP-Ribosyl-Binding and Hydrolase Activities of the Alphavirus nsP3 Macrodomain Affect Neurovirulence in Mice.mBio. 2020 Feb 11;11(1):e03253-19. doi: 10.1128/mBio.03253-19. mBio. 2020. PMID: 32047134 Free PMC article.
-
Host Factor Nucleophosmin 1 (NPM1/B23) Exerts Antiviral Effects against Chikungunya Virus by Its Interaction with Viral Nonstructural Protein 3.Microbiol Spectr. 2023 Aug 17;11(4):e0537122. doi: 10.1128/spectrum.05371-22. Epub 2023 Jul 6. Microbiol Spectr. 2023. PMID: 37409962 Free PMC article.
-
Cell-Type-Dependent Role for nsP3 Macrodomain ADP-Ribose Binding and Hydrolase Activity during Chikungunya Virus Infection.Viruses. 2022 Dec 9;14(12):2744. doi: 10.3390/v14122744. Viruses. 2022. PMID: 36560748 Free PMC article.
-
Antiviral Strategies against Arthritogenic Alphaviruses.Microorganisms. 2020 Sep 7;8(9):1365. doi: 10.3390/microorganisms8091365. Microorganisms. 2020. PMID: 32906603 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
