Recruitment of CXCR4+ type 1 innate lymphoid cells distinguishes sarcoidosis from other skin granulomatous diseases
- PMID: 39225100
- PMCID: PMC11364400
- DOI: 10.1172/JCI178711
Recruitment of CXCR4+ type 1 innate lymphoid cells distinguishes sarcoidosis from other skin granulomatous diseases
Abstract
Sarcoidosis is a multiorgan granulomatous disease that lacks diagnostic biomarkers and targeted treatments. Using blood and skin from patients with sarcoid and non-sarcoid skin granulomas, we discovered that skin granulomas from different diseases exhibit unique immune cell recruitment and molecular signatures. Sarcoid skin granulomas were specifically enriched for type 1 innate lymphoid cells (ILC1s) and B cells and exhibited molecular programs associated with formation of mature tertiary lymphoid structures (TLSs), including increased CXCL12/CXCR4 signaling. Lung sarcoidosis granulomas also displayed similar immune cell recruitment. Thus, granuloma formation was not a generic molecular response. In addition to tissue-specific effects, patients with sarcoidosis exhibited an 8-fold increase in circulating ILC1s, which correlated with treatment status. Multiple immune cell types induced CXCL12/CXCR4 signaling in sarcoidosis, including Th1 T cells, macrophages, and ILCs. Mechanistically, CXCR4 inhibition reduced sarcoidosis-activated immune cell migration, and targeting CXCR4 or total ILCs attenuated granuloma formation in a noninfectious mouse model. Taken together, our results show that ILC1s are a tissue and circulating biomarker that distinguishes sarcoidosis from other skin granulomatous diseases. Repurposing existing CXCR4 inhibitors may offer a new targeted treatment for this devastating disease.
Keywords: Cellular immune response; Dermatology; Immunology; Innate immunity.
Figures


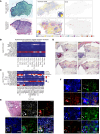


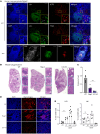
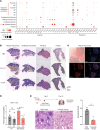
Comment in
- Type 1 innate lymphoid cells: a biomarker and therapeutic candidate in sarcoidosis
Similar articles
-
Type 1 innate lymphoid cells: a biomarker and therapeutic candidate in sarcoidosis.J Clin Invest. 2024 Sep 3;134(17):e183708. doi: 10.1172/JCI183708. J Clin Invest. 2024. PMID: 39225095 Free PMC article.
-
[Cutaneous sarcoid granuloma: cytoimmunological study of the lymphocytes (freed cells and tissue sections) (author's transl)].Ann Immunol (Paris). 1978 Jan;129(1):97-106. Ann Immunol (Paris). 1978. PMID: 655650 French.
-
Comparison of the T cell patterns in leprous and cutaneous sarcoid granulomas. Presence of Valpha24-invariant natural killer T cells in T-cell-reactive leprosy together with a highly biased T cell receptor Valpha repertoire.Am J Pathol. 2000 Aug;157(2):509-23. doi: 10.1016/s0002-9440(10)64562-2. Am J Pathol. 2000. PMID: 10934154 Free PMC article.
-
Immunopathogenesis of sarcoidosis.Clin Dermatol. 2007 May-Jun;25(3):250-8. doi: 10.1016/j.clindermatol.2007.03.002. Clin Dermatol. 2007. PMID: 17560302 Review.
-
Sarcoidosis.Dermatol Clin. 2015 Jul;33(3):389-416. doi: 10.1016/j.det.2015.03.006. Dermatol Clin. 2015. PMID: 26143421 Review.
References
-
- Judson MA, et al. The WASOG Sarcoidosis Organ Assessment Instrument: an update of a previous clinical tool. Sarcoidosis Vasc Diffuse Lung Dis. 2014;31(1):19–27. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

