Protective effects of Ganoderma lucidum spores on estradiol benzoate-induced TEC apoptosis and compromised double-positive thymocyte development
- PMID: 39221140
- PMCID: PMC11361955
- DOI: 10.3389/fphar.2024.1419881
Protective effects of Ganoderma lucidum spores on estradiol benzoate-induced TEC apoptosis and compromised double-positive thymocyte development
Abstract
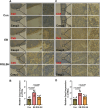
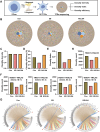
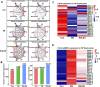
Backgroud: Thymic atrophy marks the onset of immune aging, precipitating developmental anomalies in T cells. Numerous clinical and preclinical investigations have underscored the regulatory role of Ganoderma lucidum spores (GLS) in T cell development. However, the precise mechanisms underlying this regulation remain elusive. Methods: In this study, a mice model of estradiol benzoate (EB)-induced thymic atrophy was constructed, and the improvement effect of GLS on thymic atrophy was evaluated. Then, we employs multi-omics techniques to elucidate how GLS modulates T cell development amidst EB-induced thymic atrophy in mice. Results: GLS effectively mitigates EB-induced thymic damage by attenuating apoptotic thymic epithelial cells (TECs) and enhancing the output of CD4+ T cells into peripheral blood. During thymic T cell development, sporoderm-removed GLS (RGLS) promotes T cell receptor (TCR) α rearrangement by augmenting V-J fragment rearrangement frequency and efficiency. Notably, biased Vα14-Jα18 rearrangement fosters double-positive (DP) to invariant natural killer T (iNKT) cell differentiation, partially contingent on RGLS-mediated restriction of peptide-major histocompatibility complex I (pMHCⅠ)-CD8 interaction and augmented CD1d expression in DP thymocytes, thereby promoting DP to CD4+ iNKT cell development. Furthermore, RGLS amplifies interaction between a DP subpopulation, termed DPsel-7, and plasmacytoid dendritic cells (pDCs), likely facilitating the subsequent development of double-negative iNKT1 cells. Lastly, RGLS suppresses EB-induced upregulation of Abpob and Apoa4, curbing the clearance of CD4+Abpob+ and CD4+Apoa4+ T cells by mTECs, resulting in enhanced CD4+ T cell output. Discussion: These findings indicate that the RGLS effectively mitigates EB-induced TEC apoptosis and compromised double-positive thymocyte development. These insights into RGLS's immunoregulatory role pave the way for its potential as a T-cell regeneration inducer.
Keywords: Ganoderma lucidum spores; T cell development; T cell receptor gene rearrangement; proteomics; single-cell RNA sequencing; thymic atrophy.
Copyright © 2024 Yang, Pan, Wang, Li, Zhang, Fan and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures







Similar articles
-
Comparative pharmacokinetic analysis of sporoderm-broken and sporoderm-removed Ganoderma lucidum spore in rat by using a sensitive plasma UPLC-QqQ-MS method.Biomed Chromatogr. 2024 Feb;38(2):e5787. doi: 10.1002/bmc.5787. Epub 2023 Dec 1. Biomed Chromatogr. 2024. PMID: 38038157
-
LAMP2 regulates autophagy in the thymic epithelium and thymic stroma-dependent CD4 T cell development.Autophagy. 2023 Feb;19(2):426-439. doi: 10.1080/15548627.2022.2074105. Epub 2022 May 19. Autophagy. 2023. PMID: 35535798 Free PMC article.
-
T cell factor-1 controls the lifetime of CD4+ CD8+ thymocytes in vivo and distal T cell receptor α-chain rearrangement required for NKT cell development.PLoS One. 2014 Dec 23;9(12):e115803. doi: 10.1371/journal.pone.0115803. eCollection 2014. PLoS One. 2014. PMID: 25536344 Free PMC article.
-
Thymic commitment of regulatory T cells is a pathway of TCR-dependent selection that isolates repertoires undergoing positive or negative selection.Curr Top Microbiol Immunol. 2005;293:43-71. doi: 10.1007/3-540-27702-1_3. Curr Top Microbiol Immunol. 2005. PMID: 15981475 Review.
-
Cytokines in the thymus: production and biological effects.Curr Med Chem. 2004 Feb;11(4):447-64. doi: 10.2174/0929867043455972. Curr Med Chem. 2004. PMID: 14965226 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

