Genomic and physiological properties of Anoxybacterium hadale gen. nov. sp. nov. reveal the important role of dissolved organic sulfur in microbial metabolism in hadal ecosystems
- PMID: 39220043
- PMCID: PMC11362086
- DOI: 10.3389/fmicb.2024.1423245
Genomic and physiological properties of Anoxybacterium hadale gen. nov. sp. nov. reveal the important role of dissolved organic sulfur in microbial metabolism in hadal ecosystems
Abstract
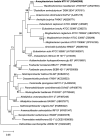
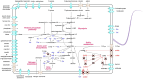
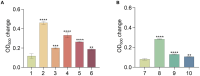
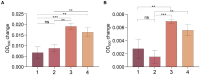
Hadal zones account for the deepest 45% of the oceanic depth range and play an important role in ocean biogeochemical cycles. As the least-explored aquatic habitat on earth, hadal ecosystems contain a vast diversity of so far uncultured microorganisms that cannot be grown on conventional laboratory culture media. Therefore, it has been difficult to gain a true understanding of the detailed metabolic characteristics and ecological functions of those difficult-to-culture microorganisms in hadal environments. In this study, a novel anaerobic bacterial strain, MT110T, was isolated from a hadal sediment-water interface sample of the Mariana Trench at 10,890 m. The level of 16S rRNA gene sequence similarity and percentage of conserved proteins between strain MT110T and the closest relatives, Anaerovorax odorimutans DSM 5092T (94.9 and 46.6%) and Aminipila butyrica DSM 103574T (94.4 and 46.7%), indicated that strain MT110T exhibits sufficient molecular differences for genus-level delineation. Phylogenetic analyses based on both 16S rRNA gene and genome sequences showed that strain MT110T formed an independent monophyletic branch within the family Anaerovoracaceae. The combined evidence showed that strain MT110T represents a novel species of a novel genus, proposed as Anoxybacterium hadale gen. nov. sp. nov. (type strain MT110T = KCTC 15922T = MCCC 1K04061T), which represents a previously uncultured lineage of the class Clostridia. Physiologically, no tested organic matter could be used as sole carbon source by strain MT110T. Genomic analysis showed that MT110T had the potential capacity of utilizing various carbon sources, but the pathways of sulfur reduction were largely incomplete. Our experiments further revealed that cysteine is one of the essential nutrients for the survival of strain MT110T, and cannot be replaced by sulfite, leucine, or taurine. This result suggests that organic sulfur compounds might play an important role in metabolism and growth of the family Anaerovoracaceae and could be one of the key factors affecting the cultivation of the uncultured microbes. Our study brings a new perspective to the role of dissolved organic sulfur in hadal ecosystems and also provides valuable information for optimizing the conditions of isolating related microbial taxa from the hadal environment.
Keywords: Clostridia bacterium; cysteine; hadal environment; organic sulfur; uncultured microbe.
Copyright © 2024 Cao, Shao, Lin, Liu, Cui, Wang and Fang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Composition and Ecological Roles of the Core Microbiome along the Abyssal-Hadal Transition Zone Sediments of the Mariana Trench.Microbiol Spectr. 2022 Jun 29;10(3):e0198821. doi: 10.1128/spectrum.01988-21. Epub 2022 Jun 7. Microbiol Spectr. 2022. PMID: 35768947 Free PMC article.
-
Identification and genomic analysis of temperate Halomonas bacteriophage vB_HmeY_H4907 from the surface sediment of the Mariana Trench at a depth of 8,900 m.Microbiol Spectr. 2023 Sep 20;11(5):e0191223. doi: 10.1128/spectrum.01912-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37728551 Free PMC article.
-
Winogradskyella ouciana sp. nov., isolated from the hadal seawater of the Mariana Trench.Int J Syst Evol Microbiol. 2019 Jun;71(3). doi: 10.1099/ijsem.0.004687. Epub 2021 Feb 8. Int J Syst Evol Microbiol. 2019. PMID: 33555246
-
Co-culture of a Novel Fermentative Bacterium, Lucifera butyrica gen. nov. sp. nov., With the Sulfur Reducer Desulfurella amilsii for Enhanced Sulfidogenesis.Front Microbiol. 2018 Dec 13;9:3108. doi: 10.3389/fmicb.2018.03108. eCollection 2018. Front Microbiol. 2018. PMID: 30631314 Free PMC article.
-
Marinobacter salinexigens sp. nov., a marine bacterium isolated from hadal seawater of the Mariana Trench.Int J Syst Evol Microbiol. 2020 Jun;70(6):3794-3800. doi: 10.1099/ijsem.0.004236. Int J Syst Evol Microbiol. 2020. PMID: 32441615
References
-
- Cao J., Gayet N., Zeng X., Shao Z., Jebbar M., Alain K. (2016). Pseudodesulfovibrio indicus gen. nov., sp. nov., a piezophilic sulfate-reducing bacterium from the Indian Ocean and reclassification of four species of the genus Desulfovibrio. Int. J. Syst. Evol. Microbiol. 66, 3904–3911. doi: 10.1099/ijsem.0.001286, PMID: - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

