TgVax452, an epitope-based candidate vaccine targeting Toxoplasma gondii tachyzoite-specific SAG1-related sequence (SRS) proteins: immunoinformatics, structural simulations and experimental evidence-based approaches
- PMID: 39210269
- PMCID: PMC11361240
- DOI: 10.1186/s12879-024-09807-x
TgVax452, an epitope-based candidate vaccine targeting Toxoplasma gondii tachyzoite-specific SAG1-related sequence (SRS) proteins: immunoinformatics, structural simulations and experimental evidence-based approaches
Erratum in
-
Correction: TgVax452, an epitope-based candidate vaccine targeting Toxoplasma gondii tachyzoite-specific SAG1-related sequence (SRS) proteins: immunoinformatics, structural simulations and experimental evidence-based approaches.BMC Infect Dis. 2024 Sep 13;24(1):969. doi: 10.1186/s12879-024-09881-1. BMC Infect Dis. 2024. PMID: 39271976 Free PMC article. No abstract available.
Abstract
Background: The highly expressed surface antigen 1 (SAG1)-related sequence (SRS) proteins of T. gondii tachyzoites, as a widespread zoonotic parasite, are critical for host cell invasion and represent promising vaccine targets. In this study, we employed a computer-aided multi-method approach for in silico design and evaluation of TgVax452, an epitope-based candidate vaccine against T. gondii tachyzoite-specific SRS proteins.
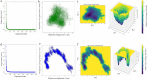
Methods: Using immunoinformatics web-based tools, structural modeling, and static/dynamic molecular simulations, we identified and screened B- and T-cell immunodominant epitopes and predicted TgVax452's antigenicity, stability, safety, adjuvanticity, and physico-chemical properties.
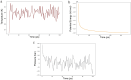
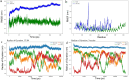
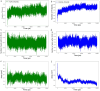
Results: The designed protein possessed 452 residues, a MW of 44.07 kDa, an alkaline pI (6.7), good stability (33.20), solubility (0.498), and antigenicity (0.9639) with no allergenicity. Comprehensive molecular dynamic (MD) simulation analyses confirmed the stable interaction (average potential energy: 3.3799 × 106 KJ/mol) between the TLR4 agonist residues (RS09 peptide) of the TgVax452 in interaction with human TLR4, potentially activating innate immune responses. Also, a dramatic increase was observed in specific antibodies (IgM and IgG), cytokines (IFN-γ), and lymphocyte responses, based on C-ImmSim outputs. Finally, we optimized TgVax452's codon adaptation and mRNA secondary structure for efficient expression in E. coli BL21 expression machinery.
Conclusion: Our findings suggest that TgVax452 is a promising candidate vaccine against T. gondii tachyzoite-specific SRS proteins and requires further experimental studies for its potential use in preclinical trials.
Keywords: Toxoplasma Gondii; Dynamic simulations; SRS proteins.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
In silico analysis and expression of a novel chimeric antigen as a vaccine candidate against Toxoplasma gondii.Microb Pathog. 2019 Jul;132:275-281. doi: 10.1016/j.micpath.2019.05.013. Epub 2019 May 9. Microb Pathog. 2019. PMID: 31078709
-
Toxoplasma gondii: Vaccination with a DNA vaccine encoding T- and B-cell epitopes of SAG1, GRA2, GRA7 and ROP16 elicits protection against acute toxoplasmosis in mice.Vaccine. 2015 Nov 27;33(48):6757-62. doi: 10.1016/j.vaccine.2015.10.077. Epub 2015 Oct 27. Vaccine. 2015. PMID: 26518401
-
A4D12 monoclonal antibody recognizes a new linear epitope from SAG2A Toxoplasma gondii tachyzoites, identified by phage display bioselection.Immunobiology. 2010;215(1):26-37. doi: 10.1016/j.imbio.2009.01.008. Epub 2009 Mar 3. Immunobiology. 2010. PMID: 19261354
-
Vaccination with a novel multi-epitope ROP8 DNA vaccine against acute Toxoplasma gondii infection induces strong B and T cell responses in mice.Comp Immunol Microbiol Infect Dis. 2020 Apr;69:101413. doi: 10.1016/j.cimid.2020.101413. Epub 2020 Jan 8. Comp Immunol Microbiol Infect Dis. 2020. PMID: 31954995
-
Candidate antigenic epitopes for vaccination and diagnosis strategies of Toxoplasma gondii infection: A review.Microb Pathog. 2019 Dec;137:103788. doi: 10.1016/j.micpath.2019.103788. Epub 2019 Oct 9. Microb Pathog. 2019. PMID: 31605758 Review.
Cited by
-
Correction: TgVax452, an epitope-based candidate vaccine targeting Toxoplasma gondii tachyzoite-specific SAG1-related sequence (SRS) proteins: immunoinformatics, structural simulations and experimental evidence-based approaches.BMC Infect Dis. 2024 Sep 13;24(1):969. doi: 10.1186/s12879-024-09881-1. BMC Infect Dis. 2024. PMID: 39271976 Free PMC article. No abstract available.
References
-
- Amouei A, Sarvi S, Sharif M, Aghayan SA, Javidnia J, Mizani A, et al. A systematic review of Toxoplasma gondii genotypes and feline: geographical distribution trends. Transbound Emerg Dis. 2020;67(1):46–64. - PubMed
-
- Fallahi S, Rostami A, Shiadeh MN, Behniafar H, Paktinat S. An updated literature review on maternal-fetal and reproductive disorders of Toxoplasma Gondii infection. J Gynecol Obstet Hum Reprod. 2018;47(3):133–40. - PubMed
-
- Foroutan-Rad M, Majidiani H, Dalvand S, Daryani A, Kooti W, Saki J, et al. Toxoplasmosis in blood donors: a systematic review and meta-analysis. Transfus Med Rev. 2016;30(3):116–22. - PubMed
-
- Dard C, Marty P, Brenier-Pinchart M-P, Garnaud C, Fricker-Hidalgo H, Pelloux H, et al. Management of toxoplasmosis in transplant recipients: an update. Expert Rev Anti-Infective Therapy. 2018;16(6):447–60. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

