PRDM16-DT is a novel lncRNA that regulates astrocyte function in Alzheimer's disease
- PMID: 39207536
- PMCID: PMC11362476
- DOI: 10.1007/s00401-024-02787-x
PRDM16-DT is a novel lncRNA that regulates astrocyte function in Alzheimer's disease
Abstract
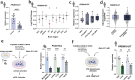
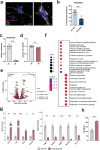
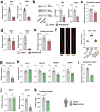
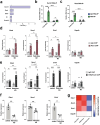
Astrocytes provide crucial support for neurons, contributing to synaptogenesis, synaptic maintenance, and neurotransmitter recycling. Under pathological conditions, deregulation of astrocytes contributes to neurodegenerative diseases such as Alzheimer's disease (AD). While most research in this field has focused on protein-coding genes, non-coding RNAs, particularly long non-coding RNAs (lncRNAs), have emerged as significant regulatory molecules. In this study, we identified the lncRNA PRDM16-DT as highly enriched in the human brain, where it is almost exclusively expressed in astrocytes. PRDM16-DT and its murine homolog, Prdm16os, are downregulated in the brains of AD patients and in AD models. In line with this, knockdown of PRDM16-DT and Prdm16os revealed its critical role in maintaining astrocyte homeostasis and supporting neuronal function by regulating genes essential for glutamate uptake, lactate release, and neuronal spine density through interactions with the RE1-Silencing Transcription factor (Rest) and Polycomb Repressive Complex 2 (PRC2). Notably, CRISPR-mediated overexpression of Prdm16os mitigated functional deficits in astrocytes induced by stimuli linked to AD pathogenesis. These findings underscore the importance of PRDM16-DT in astrocyte function and its potential as a novel therapeutic target for neurodegenerative disorders characterized by astrocyte dysfunction.
Keywords: Alzheimer’s disease; Astrocyte; Brain; Long-non-coding RNA; Postmortem human brain.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
PRDM16-DT: A Brain and Astrocyte-Specific lncRNA Implicated in Alzheimer's Disease.bioRxiv [Preprint]. 2024 Jul 1:2024.06.27.600964. doi: 10.1101/2024.06.27.600964. bioRxiv. 2024. PMID: 39005272 Free PMC article. Preprint.
-
LINC00982-encoded protein PRDM16-DT regulates CHEK2 splicing to suppress colorectal cancer metastasis and chemoresistance.Theranostics. 2024 May 27;14(8):3317-3338. doi: 10.7150/thno.95485. eCollection 2024. Theranostics. 2024. PMID: 38855188 Free PMC article.
-
Upregulation of the lncRNA MEG3 improves cognitive impairment, alleviates neuronal damage, and inhibits activation of astrocytes in hippocampus tissues in Alzheimer's disease through inactivating the PI3K/Akt signaling pathway.J Cell Biochem. 2019 Oct;120(10):18053-18065. doi: 10.1002/jcb.29108. Epub 2019 Jun 12. J Cell Biochem. 2019. PMID: 31190362
-
Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases.Brain Res Bull. 2013 Aug;97:69-80. doi: 10.1016/j.brainresbull.2013.06.001. Epub 2013 Jun 10. Brain Res Bull. 2013. PMID: 23756188 Review.
-
Dysregulation of Astrocyte-Neuronal Communication in Alzheimer's Disease.Int J Mol Sci. 2021 Jul 23;22(15):7887. doi: 10.3390/ijms22157887. Int J Mol Sci. 2021. PMID: 34360652 Free PMC article. Review.
Cited by
-
The Association and Prognostic Implications of Long Non-Coding RNAs in Major Psychiatric Disorders, Alzheimer's Diseases and Parkinson's Diseases: A Systematic Review.Int J Mol Sci. 2024 Oct 12;25(20):10995. doi: 10.3390/ijms252010995. Int J Mol Sci. 2024. PMID: 39456775 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

