The Dual Role of TRIM7 in Viral Infections
- PMID: 39205259
- PMCID: PMC11360163
- DOI: 10.3390/v16081285
The Dual Role of TRIM7 in Viral Infections
Abstract
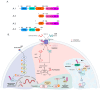
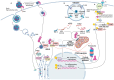
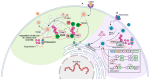
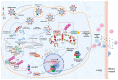
The E3 ubiquitin ligase TRIM7 is known to have dual roles during viral infections. Like other TRIM proteins, TRIM7 can regulate the IFN pathway via the regulation of the cytosolic receptors RIG-I or MDA-5, which promote the production of type I interferons (IFN-I) and antiviral immune responses. Alternatively, under certain infectious conditions, TRIM7 can negatively regulate IFN-I signaling, resulting in increased virus replication. A growing body of evidence has also shown that TRIM7 can, in some cases, ubiquitinate viral proteins to promote viral replication and pathogenesis, while in other cases it can promote degradation of viral proteins through the proteasome, reducing virus infection. TRIM7 can also regulate the host inflammatory response and modulate the production of inflammatory cytokines, which can lead to detrimental inflammation. TRIM7 can also protect the host during infection by reducing cellular apoptosis. Here, we discuss the multiple functions of TRIM7 during viral infections and its potential as a therapeutic target.
Keywords: TRIM7; antiviral response; ubiquitination; viral infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
To TRIM or not to TRIM: the balance of host-virus interactions mediated by the ubiquitin system.J Gen Virol. 2019 Dec;100(12):1641-1662. doi: 10.1099/jgv.0.001341. J Gen Virol. 2019. PMID: 31661051 Free PMC article.
-
TRIM7 inhibits enterovirus replication and promotes emergence of a viral variant with increased pathogenicity.Cell. 2021 Jun 24;184(13):3410-3425.e17. doi: 10.1016/j.cell.2021.04.047. Epub 2021 May 31. Cell. 2021. PMID: 34062120 Free PMC article.
-
The Host E3-Ubiquitin Ligase TRIM6 Ubiquitinates the Ebola Virus VP35 Protein and Promotes Virus Replication.J Virol. 2017 Aug 24;91(18):e00833-17. doi: 10.1128/JVI.00833-17. Print 2017 Sep 15. J Virol. 2017. PMID: 28679761 Free PMC article.
-
Interplay between TRIM7 and antiviral immunity.Front Cell Infect Microbiol. 2023 Aug 31;13:1256882. doi: 10.3389/fcimb.2023.1256882. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37719674 Free PMC article. Review.
-
Tripartite motif-containing proteins precisely and positively affect host antiviral immune response.Scand J Immunol. 2018 Jun;87(6):e12669. doi: 10.1111/sji.12669. Scand J Immunol. 2018. PMID: 29706026 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

