VOGDB-Database of Virus Orthologous Groups
- PMID: 39205165
- PMCID: PMC11360334
- DOI: 10.3390/v16081191
VOGDB-Database of Virus Orthologous Groups
Abstract
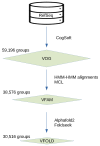
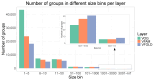
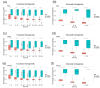
Computational models of homologous protein groups are essential in sequence bioinformatics. Due to the diversity and rapid evolution of viruses, the grouping of protein sequences from virus genomes is particularly challenging. The low sequence similarities of homologous genes in viruses require specific approaches for sequence- and structure-based clustering. Furthermore, the annotation of virus genomes in public databases is not as consistent and up to date as for many cellular genomes. To tackle these problems, we have developed VOGDB, which is a database of virus orthologous groups. VOGDB is a multi-layer database that progressively groups viral genes into groups connected by increasingly remote similarity. The first layer is based on pair-wise sequence similarities, the second layer is based on the sequence profile alignments, and the third layer uses predicted protein structures to find the most remote similarity. VOGDB groups allow for more sensitive homology searches of novel genes and increase the chance of predicting annotations or inferring phylogeny. VOGD B uses all virus genomes from RefSeq and partially reannotates them. VOGDB is updated with every RefSeq release. The unique feature of VOGDB is the inclusion of both prokaryotic and eukaryotic viruses in the same clustering process, which makes it possible to explore old evolutionary relationships of the two groups. VOGDB is freely available at vogdb.org under the CC BY 4.0 license.
Keywords: comparative Genomics; genome analysis; genome annotation; orthologous groups; protein families; virus genomes.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Prokaryotic Virus Orthologous Groups (pVOGs): a resource for comparative genomics and protein family annotation.Nucleic Acids Res. 2017 Jan 4;45(D1):D491-D498. doi: 10.1093/nar/gkw975. Epub 2016 Oct 26. Nucleic Acids Res. 2017. PMID: 27789703 Free PMC article.
-
PhEVER: a database for the global exploration of virus-host evolutionary relationships.Nucleic Acids Res. 2011 Jan;39(Database issue):D569-75. doi: 10.1093/nar/gkq1013. Epub 2010 Nov 16. Nucleic Acids Res. 2011. PMID: 21081560 Free PMC article.
-
Bioinformatic Approaches for Comparative Analysis of Viruses.Methods Mol Biol. 2018;1704:401-417. doi: 10.1007/978-1-4939-7463-4_15. Methods Mol Biol. 2018. PMID: 29277875
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review.
References
-
- Villarreal L. Encyclopedia of Virology. Elsevier; Amsterdam, The Netherlands: 2008. Evolution of Viruses; pp. 174–184. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

