Microbial Metagenomics Revealed the Diversity and Distribution Characteristics of Groundwater Microorganisms in the Middle and Lower Reaches of the Yangtze River Basin
- PMID: 39203393
- PMCID: PMC11356026
- DOI: 10.3390/microorganisms12081551
Microbial Metagenomics Revealed the Diversity and Distribution Characteristics of Groundwater Microorganisms in the Middle and Lower Reaches of the Yangtze River Basin
Abstract
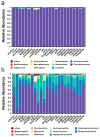
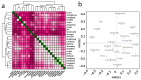
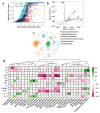
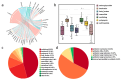
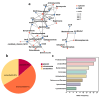
Groundwater is one of the important freshwater resources on Earth and is closely related to human activities. As a good biological vector, a more diverse repertory of antibiotic resistance genes in the water environment would have a profound impact on human medical health. Therefore, this study conducted a metagenomic sequencing analysis of water samples from groundwater monitoring points in the middle and lower reaches of the Yangtze River to characterize microbial community composition and antibiotic resistance in the groundwater environment. Our results show that different microbial communities and community composition were the driving factors in the groundwater environment, and a diversity of antibiotic resistance genes in the groundwater environment was detected. The main source of antibiotic resistance gene host was determined by correlation tests and analyses. In this study, metagenomics was used for the first time to comprehensively analyze microbial communities in groundwater systems in the middle and lower reaches of the Yangtze River basin. The data obtained from this study serve as an invaluable resource and represent the basic metagenomic characteristics of groundwater microbial communities in the middle and lower reaches of the Yangtze River basin. These findings will be useful tools and provide a basis for future research on water microbial community and quality, greatly expanding the depth and breadth of our understanding of groundwater.
Keywords: antibiotic-resistant bacteria; diversity; groundwater; metagenomics; microorganisms.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
[Distributions of Antibiotic Resistance Genes and Microbial Communities in the Nearshore Area of the Yangtze River Estuary].Huan Jing Ke Xue. 2023 Jan 8;44(1):158-168. doi: 10.13227/j.hjkx.202203160. Huan Jing Ke Xue. 2023. PMID: 36635804 Chinese.
-
Spatial and temporal distribution and affecting factors of iron and manganese in the groundwater in the middle area of the Yangtze River Basin, China.Environ Sci Pollut Res Int. 2022 Aug;29(40):61204-61221. doi: 10.1007/s11356-022-20253-7. Epub 2022 Apr 19. Environ Sci Pollut Res Int. 2022. PMID: 35441292
-
[Spatiotemporal Variation Characteristics of Main Pollutant Fluxes in the Yangtze River Basin from 2017 to 2020].Huan Jing Ke Xue. 2023 Aug 8;44(8):4279-4291. doi: 10.13227/j.hjkx.202209245. Huan Jing Ke Xue. 2023. PMID: 37694623 Chinese.
-
[Community Composition and Assessment of the Aquatic Ecosystem of Periphytic Algae in the Yangtze River Basin].Huan Jing Ke Xue. 2022 Aug 8;43(8):3998-4007. doi: 10.13227/j.hjkx.202111229. Huan Jing Ke Xue. 2022. PMID: 35971698 Chinese.
-
Distribution, formation and human-induced evolution of geogenic contaminated groundwater in China: A review.Sci Total Environ. 2018 Dec 1;643:967-993. doi: 10.1016/j.scitotenv.2018.06.201. Epub 2018 Jun 28. Sci Total Environ. 2018. PMID: 29960233 Review.
References
-
- Azam F. Microbial control of oceanic carbon flux: The plot thickens. Science. 1998;280:694–696. doi: 10.1126/science.280.5364.694. - DOI
-
- Gleick P.H. Water in Crisis: A Guide to the World’s Fresh Water Resources. Oxford University Press; New York, NY, USA: 1993.
-
- Griebler C., Lueders T. Microbial biodiversity in groundwater ecosystems. Freshwater Biol. 2009;54:649–677. doi: 10.1111/j.1365-2427.2008.02013.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

