Molecular Signatures of CB-6644 Inhibition of the RUVBL1/2 Complex in Multiple Myeloma
- PMID: 39201707
- PMCID: PMC11354775
- DOI: 10.3390/ijms25169022
Molecular Signatures of CB-6644 Inhibition of the RUVBL1/2 Complex in Multiple Myeloma
Abstract
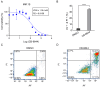
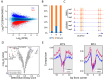
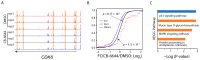
Multiple myeloma is the second most hematological cancer. RUVBL1 and RUVBL2 form a subcomplex of many chromatin remodeling complexes implicated in cancer progression. As an inhibitor specific to the RUVBL1/2 complex, CB-6644 exhibits remarkable anti-tumor activity in xenograft models of Burkitt's lymphoma and multiple myeloma (MM). In this work, we defined transcriptional signatures corresponding to CB-6644 treatment in MM cells and determined underlying epigenetic changes in terms of chromatin accessibility. CB-6644 upregulated biological processes related to interferon response and downregulated those linked to cell proliferation in MM cells. Transcriptional regulator inference identified E2Fs as regulators for downregulated genes and MED1 and MYC as regulators for upregulated genes. CB-6644-induced changes in chromatin accessibility occurred mostly in non-promoter regions. Footprinting analysis identified transcription factors implied in modulating chromatin accessibility in response to CB-6644 treatment, including ATF4/CEBP and IRF4. Lastly, integrative analysis of transcription responses to various chemical compounds of the molecular signature genes from public gene expression data identified CB-5083, a p97 inhibitor, as a synergistic candidate with CB-6644 in MM cells, but experimental validation refuted this hypothesis.
Keywords: CB-6644; RUVBL1/2; molecular signatures; multiple myeloma.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
CB-6644 Is a Selective Inhibitor of the RUVBL1/2 Complex with Anticancer Activity.ACS Chem Biol. 2019 Feb 15;14(2):236-244. doi: 10.1021/acschembio.8b00904. Epub 2019 Jan 25. ACS Chem Biol. 2019. PMID: 30640450
-
CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling.Mol Cancer. 2019 Oct 29;18(1):150. doi: 10.1186/s12943-019-1076-1. Mol Cancer. 2019. PMID: 31665067 Free PMC article.
-
RUVBL1 is an amplified epigenetic factor promoting proliferation and inhibiting differentiation program in head and neck squamous cancers.Oral Oncol. 2020 Dec;111:104930. doi: 10.1016/j.oraloncology.2020.104930. Epub 2020 Jul 31. Oral Oncol. 2020. PMID: 32745900
-
RUVBL1-RUVBL2 AAA-ATPase: a versatile scaffold for multiple complexes and functions.Curr Opin Struct Biol. 2021 Apr;67:78-85. doi: 10.1016/j.sbi.2020.08.010. Epub 2020 Oct 28. Curr Opin Struct Biol. 2021. PMID: 33129013 Review.
-
Epigenetic regulatory mutations and epigenetic therapy for multiple myeloma.Curr Opin Hematol. 2017 Jul;24(4):336-344. doi: 10.1097/MOH.0000000000000358. Curr Opin Hematol. 2017. PMID: 28441149 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

