NK Cell Degranulation Triggered by Rituximab Identifies Potential Markers of Subpopulations with Enhanced Cytotoxicity toward Malignant B Cells
- PMID: 39201666
- PMCID: PMC11354239
- DOI: 10.3390/ijms25168980
NK Cell Degranulation Triggered by Rituximab Identifies Potential Markers of Subpopulations with Enhanced Cytotoxicity toward Malignant B Cells
Abstract
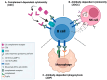
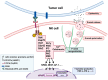
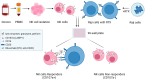
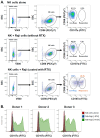
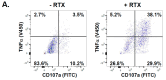
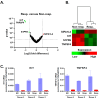
A promising strategy in cancer immunotherapy is to restore or enhance the cytotoxicity of NK cells, among others, by activating the mechanism of antibody-dependent cellular cytotoxicity (ADCC). Monoclonal antibodies targeting tumor antigens, such as rituximab (targeting CD20), induce NK cell-mediated ADCC and have been used to treat B cell malignancies, such as non-Hodgkin lymphoma, but not always successfully. The aim of this study was to analyze the gene expression profile of the NK cells involved in the cytolytic response stimulated by rituximab. NK cells were co-cultured with rituximab-opsonized Raji cells. Sorting into responder and non-responder groups was based on the presence of CD107a, which is a degranulation marker. RNA-seq results showed that the KIT and TNFSF4 genes were strongly down-regulated in the degranulating population of NK cells (responders); this was further confirmed by qRT-PCR. Both genes encode surface proteins with cellular signaling abilities, namely c-KIT and the OX40 ligand. Consistent with our findings, c-KIT was previously reported to correlate inversely with cytokine production by activated NK cells. The significance of these findings for cancer immunotherapy seems essential, as the pharmacological inhibition of c-KIT and OX40L, or gene ablation, could be further tested for the enhancement of the anti-tumor activity of NK cells in response to rituximab.
Keywords: Burkitt lymphoma; CD16; NK cells; OX40L; c-KIT; mAbs; rituximab.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
The anti-lymphoma effect of antibody-mediated immunotherapy is based on an increased degranulation of peripheral blood natural killer (NK) cells.Exp Hematol. 2006 Jun;34(6):753-9. doi: 10.1016/j.exphem.2006.02.015. Exp Hematol. 2006. PMID: 16728280
-
Expansion of Human NK Cells Using K562 Cells Expressing OX40 Ligand and Short Exposure to IL-21.Front Immunol. 2019 Apr 24;10:879. doi: 10.3389/fimmu.2019.00879. eCollection 2019. Front Immunol. 2019. PMID: 31105701 Free PMC article.
-
Non-Coated Rituximab Induces Highly Cytotoxic Natural Killer Cells From Peripheral Blood Mononuclear Cells via Autologous B Cells.Front Immunol. 2021 May 25;12:658562. doi: 10.3389/fimmu.2021.658562. eCollection 2021. Front Immunol. 2021. PMID: 34113342 Free PMC article.
-
NK cells stimulated with IL-15 or CpG ODN enhance rituximab-dependent cellular cytotoxicity against B-cell lymphoma.Exp Hematol. 2008 Jan;36(1):69-77. doi: 10.1016/j.exphem.2007.08.012. Epub 2007 Oct 23. Exp Hematol. 2008. PMID: 17959301
-
A Tridimensional Model for NK Cell-Mediated ADCC of Follicular Lymphoma.Front Immunol. 2019 Aug 14;10:1943. doi: 10.3389/fimmu.2019.01943. eCollection 2019. Front Immunol. 2019. PMID: 31475004 Free PMC article.
References
-
- Veuillen C., Aurran-Schleinitz T., Castellano R., Rey J., Mallet F., Orlanducci F., Pouyet L., Just-Landi S., Coso D., Ivanov V., et al. Primary B-CLL resistance to NK cell cytotoxicity can be overcome in vitro and in vivo by priming NK cells and monoclonal antibody therapy. J. Clin. Immunol. 2012;32:632–646. doi: 10.1007/s10875-011-9624-5. - DOI - PubMed
-
- Prica A., Baldassarre F., Hicks L.K., Imrie K., Kouroukis T., Cheung M., Members of the Hematology Disease Site Group of the Cancer Care Ontario Program in Evidence-Based Care Rituximab in Lymphoma and Chronic Lymphocytic Leukaemia: A Practice Guideline. Clin. Oncol. 2017;29:e13–e28. doi: 10.1016/j.clon.2016.09.004. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

