SKF-96365 Expels Tyrosine Kinase Inhibitor-Treated CML Stem and Progenitor Cells from the HS27A Stromal Cell Niche in a RhoA-Dependent Mechanism
- PMID: 39199564
- PMCID: PMC11352811
- DOI: 10.3390/cancers16162791
SKF-96365 Expels Tyrosine Kinase Inhibitor-Treated CML Stem and Progenitor Cells from the HS27A Stromal Cell Niche in a RhoA-Dependent Mechanism
Abstract
Background: A major issue in Chronic Myeloid Leukemia (CML) is the persistence of quiescent leukemia stem cells (LSCs) in the hematopoietic niche under tyrosine kinase inhibitor (TKI) treatment.
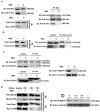
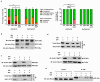
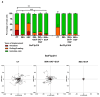
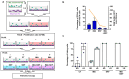
Results: Here, using CFSE sorting, we show that low-proliferating CD34+ cells from CML patients in 3D co-culture hide under HS27A stromal cells during TKI treatment-a behavior less observed in untreated cells. Under the same conditions, Ba/F3p210 cells lose their spontaneous motility. In CML CD34+ and Ba/F3p210 cells, while Rac1 is completely inhibited by TKI, RhoA remains activated but is unable to signal to ROCK. Co-incubation of Ba/F3p210 cells with TKI, SKF-96365 (a calcium channel inhibitor), and EGF restores myosin II activation and amoeboid motility to levels comparable to untreated cells, sustaining the activation of ROCK. In CFSE+ CD34+ cells containing quiescent leukemic stem cells, co-incubation of TKI with SKF-96365 induced the expulsion of these cells from the HS27A niche.
Conclusions: This study underscores the role of RhoA in LSC behavior under TKI treatment and suggests that SKF-96365 could remobilize quiescent CML LSCs through reactivation of the RhoA/ROCK pathway.
Keywords: Chronic Myeloid Leukemia; RhoGTPases; SKF-96365; leukemia niche; leukemia stem cells; tyrosine kinase inhibitors.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Overcoming BCR::ABL1 dependent and independent survival mechanisms in chronic myeloid leukaemia using a multi-kinase targeting approach.Cell Commun Signal. 2023 Nov 29;21(1):342. doi: 10.1186/s12964-023-01363-2. Cell Commun Signal. 2023. PMID: 38031192 Free PMC article.
-
Repurposing the Bis-Biguanide Alexidine in Combination with Tyrosine Kinase Inhibitors to Eliminate Leukemic Stem/Progenitor Cells in Chronic Myeloid Leukemia.Cancers (Basel). 2023 Feb 3;15(3):995. doi: 10.3390/cancers15030995. Cancers (Basel). 2023. PMID: 36765952 Free PMC article.
-
Infliximab therapy together with tyrosine kinase inhibition targets leukemic stem cells in chronic myeloid leukemia.BMC Cancer. 2019 Jul 4;19(1):658. doi: 10.1186/s12885-019-5871-2. BMC Cancer. 2019. PMID: 31272418 Free PMC article.
-
Targeting BMP signaling in the bone marrow microenvironment of myeloid leukemia.Biochem Soc Trans. 2020 Apr 29;48(2):411-418. doi: 10.1042/BST20190223. Biochem Soc Trans. 2020. PMID: 32167132 Review.
-
Leukemia Stem Cells as a Potential Target to Achieve Therapy-Free Remission in Chronic Myeloid Leukemia.Cancers (Basel). 2021 Nov 20;13(22):5822. doi: 10.3390/cancers13225822. Cancers (Basel). 2021. PMID: 34830976 Free PMC article. Review.
References
-
- Mahon F.X., Réa D., Guilhot J., Guilhot F., Huguet F., Nicolini F., Legros L., Charbonnier A., Guerci A., Varet B. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: The prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11:1029–1035. doi: 10.1016/S1470-2045(10)70233-3. - DOI - PubMed
-
- Chomel J.C., Bonnet M.L., Sorel N., Bertrand A., Meunier M.C., Fichelson S., Melkus M., Bennaceur-Griscelli A., Guilhot F., Turhan A.G. Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood. 2011;118:3657–3660. doi: 10.1182/blood-2011-02-335497. - DOI - PMC - PubMed
-
- Charaf L., Mahon F.X., Lamrissi-Garcia I., Moranvillier I., Beliveau F., Cardinaud B., Dabernat S., de Verneuil H., Moreau-Gaudry F., Bedel A. Effect of tyrosine kinase inhibitors on stemness in normal and chronic myeloid leukemia cells. Leukemia. 2017;31:65–74. doi: 10.1038/leu.2016.154. - DOI - PubMed
-
- Moreno-Lorenzana D., Avilés-Vazquez S., Sandoval Esquivel M.A., Alvarado-Moreno A., Ortiz-Navarrete V., Torres-Martínez H., Ayala-Sánchez M., Mayani H., Chavez-Gonzalez A. CDKIs p18INK4c and p57Kip2 are involved in quiescence of CML leukemic stem cells after treatment with TKI. Cell Cycle. 2016;15:1276–1287. doi: 10.1080/15384101.2016.1160976. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

