Reverse vaccinology approaches to design a potent multiepitope vaccine against the HIV whole genome: immunoinformatic, bioinformatics, and molecular dynamics approaches
- PMID: 39198721
- PMCID: PMC11360854
- DOI: 10.1186/s12879-024-09775-2
Reverse vaccinology approaches to design a potent multiepitope vaccine against the HIV whole genome: immunoinformatic, bioinformatics, and molecular dynamics approaches
Erratum in
-
Correction: Reverse vaccinology approaches to design a potent multiepitope vaccine against the HIV whole genome: immunoinformatic, bioinformatics, and molecular dynamics approaches.BMC Infect Dis. 2024 Sep 20;24(1):1023. doi: 10.1186/s12879-024-09951-4. BMC Infect Dis. 2024. PMID: 39304818 Free PMC article. No abstract available.
Abstract
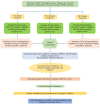
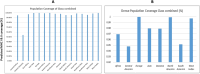
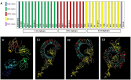
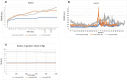
Substantial advances have been made in the development of promising HIV vaccines to eliminate HIV-1 infection. For the first time, one hundred of the most submitted HIV subtypes and CRFs were retrieved from the LANL database, and the consensus sequences of the eleven HIV proteins were obtained to design vaccines for human and mouse hosts. By using various servers and filters, highly qualified B-cell epitopes, as well as HTL and CD8 + epitopes that were common between mouse and human alleles and were also located in the conserved domains of HIV proteins, were considered in the vaccine constructs. With 90% coverage worldwide, the human vaccine model covers a diverse allelic population, making it widely available. Codon optimization and in silico cloning in prokaryotic and eukaryotic vectors guarantee high expression of the vaccine models in human and E. coli hosts. Molecular dynamics confirmed the stable interaction of the vaccine constructs with TLR3, TLR4, and TLR9, leading to a substantial immunogenic response to the designed vaccine. Vaccine models effectively target the humoral and cellular immune systems in humans and mice; however, experimental validation is needed to confirm these findings in silico.
Keywords: HIV; Bioinformatic; Main HIV subtypes and CRF; Molecular dynamic; TLR; Vaccine.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Exploring T & B-cell epitopes and designing multi-epitope subunit vaccine targeting integration step of HIV-1 lifecycle using immunoinformatics approach.Microb Pathog. 2019 Dec;137:103791. doi: 10.1016/j.micpath.2019.103791. Epub 2019 Oct 10. Microb Pathog. 2019. PMID: 31606417
-
Design of multivalent-epitope vaccine models directed toward the world's population against HIV-Gag polyprotein: Reverse vaccinology and immunoinformatics.PLoS One. 2024 Sep 27;19(9):e0306559. doi: 10.1371/journal.pone.0306559. eCollection 2024. PLoS One. 2024. PMID: 39331650 Free PMC article.
-
Immunoinformatics design of a novel epitope-based vaccine candidate against dengue virus.Sci Rep. 2021 Oct 5;11(1):19707. doi: 10.1038/s41598-021-99227-7. Sci Rep. 2021. PMID: 34611250 Free PMC article.
-
Reverse vaccinology-based multi-epitope vaccine design against Indian group A rotavirus targeting VP7, VP4, and VP6 proteins.Microb Pathog. 2024 Aug;193:106775. doi: 10.1016/j.micpath.2024.106775. Epub 2024 Jul 1. Microb Pathog. 2024. PMID: 38960216 Review.
-
Multiple Approaches for Increasing the Immunogenicity of an Epitope-Based Anti-HIV Vaccine.AIDS Res Hum Retroviruses. 2015 Nov;31(11):1077-88. doi: 10.1089/AID.2015.0101. Epub 2015 Aug 13. AIDS Res Hum Retroviruses. 2015. PMID: 26149745 Review.
Cited by
-
Correction: Reverse vaccinology approaches to design a potent multiepitope vaccine against the HIV whole genome: immunoinformatic, bioinformatics, and molecular dynamics approaches.BMC Infect Dis. 2024 Sep 20;24(1):1023. doi: 10.1186/s12879-024-09951-4. BMC Infect Dis. 2024. PMID: 39304818 Free PMC article. No abstract available.
References
-
- Hashempour A, Moayedi J, Musavi Z, Ghasabi F, Halaji M, Hasanshahi Z, Nazarinia MA. First report of HHV-8 viral load and seroprevalence of major blood-borne viruses in Iranian patients with systemic sclerosis. Multiple Scler Relat Disorders. 2021;51:102872. - PubMed
-
- Hashempour T, Bamdad T, Bergamini A, Lavergne JP, Haj-Sheykholeslami A, Brakier-Gingras L, Ajorloo M, Merat S. F protein increases CD4 + CD25 + T cell population in patients with chronic hepatitis C. Pathogens Disease. 2015;73(4):ftv022. - PubMed
-
- Halaji M, Hashempour T, Moayedi J, Pouladfar GR, Khansarinejad B, Khashei R, Moattari A, Musavi Z, Ghassabi F, Pirbonyeh N. Viral etiology of acute respiratory infections in children in Southern Iran. Rev Soc Bras Med Trop 2019, 52. - PubMed
-
- Hashempoor T, Alborzi AM, Moayedi J, Ajorloo M, Bamdad T, Sharifi AH, Lavergne JP, Haj-sheykholeslami A, Merat S. A decline in anti-core + 1 antibody titer occurs in successful treatment of patients infected with hepatitis C virus. Jundishapur J Microbiol 2018, 11(2).
-
- Dehghani B, Hashempour T, Hasanshahi Z. Interaction of human herpesvirus 8 viral interleukin-6 with human interleukin-6 receptor using in silico approach: the potential role in HHV-8 pathogenesis. Curr Proteomics. 2020;17(2):107–16.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

