Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch
- PMID: 39196936
- PMCID: PMC11352909
- DOI: 10.1126/sciadv.adp0975
Cancer-associated DNA hypermethylation of Polycomb targets requires DNMT3A dual recognition of histone H2AK119 ubiquitination and the nucleosome acidic patch
Abstract
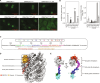
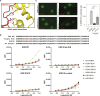
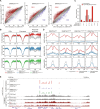
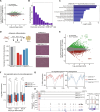
During tumor development, promoter CpG islands that are normally silenced by Polycomb repressive complexes (PRCs) become DNA-hypermethylated. The molecular mechanism by which de novo DNA methyltransferase(s) [DNMT(s)] catalyze CpG methylation at PRC-regulated regions remains unclear. Here, we report a cryo-electron microscopy structure of the DNMT3A long isoform (DNMT3A1) amino-terminal region in complex with a nucleosome carrying PRC1-mediated histone H2A lysine-119 monoubiquitination (H2AK119Ub). We identify regions within the DNMT3A1 amino terminus that bind H2AK119Ub and the nucleosome acidic patch. This bidentate interaction is required for effective DNMT3A1 engagement with H2AK119Ub-modified chromatin in cells. Further, aberrant redistribution of DNMT3A1 to Polycomb target genes recapitulates the cancer-associated DNA hypermethylation signature and inhibits their transcriptional activation during cell differentiation. This effect is rescued by disruption of the DNMT3A1-acidic patch interaction. Together, our analyses reveal a binding interface critical for mediating promoter CpG island DNA hypermethylation, a major molecular hallmark of cancer.
Figures

Update of
-
Cancer-associated DNA Hypermethylation of Polycomb Targets Requires DNMT3A Dual Recognition of Histone H2AK119 Ubiquitination and the Nucleosome Acidic Patch.bioRxiv [Preprint]. 2024 Mar 20:2024.03.18.585588. doi: 10.1101/2024.03.18.585588. bioRxiv. 2024. Update in: Sci Adv. 2024 Aug 30;10(35):eadp0975. doi: 10.1126/sciadv.adp0975 PMID: 38562823 Free PMC article. Updated. Preprint.
Similar articles
-
Cancer-associated DNA Hypermethylation of Polycomb Targets Requires DNMT3A Dual Recognition of Histone H2AK119 Ubiquitination and the Nucleosome Acidic Patch.bioRxiv [Preprint]. 2024 Mar 20:2024.03.18.585588. doi: 10.1101/2024.03.18.585588. bioRxiv. 2024. Update in: Sci Adv. 2024 Aug 30;10(35):eadp0975. doi: 10.1126/sciadv.adp0975 PMID: 38562823 Free PMC article. Updated. Preprint.
-
Two competing mechanisms of DNMT3A recruitment regulate the dynamics of de novo DNA methylation at PRC1-targeted CpG islands.Nat Genet. 2021 Jun;53(6):794-800. doi: 10.1038/s41588-021-00856-5. Epub 2021 May 13. Nat Genet. 2021. PMID: 33986537 Free PMC article.
-
Structural basis for the H2AK119ub1-specific DNMT3A-nucleosome interaction.Nat Commun. 2024 Jul 23;15(1):6217. doi: 10.1038/s41467-024-50526-3. Nat Commun. 2024. PMID: 39043678 Free PMC article.
-
Activity of PRC1 and Histone H2AK119 Monoubiquitination: Revising Popular Misconceptions.Bioessays. 2020 May;42(5):e1900192. doi: 10.1002/bies.201900192. Epub 2020 Mar 20. Bioessays. 2020. PMID: 32196702 Free PMC article. Review.
-
Polycomb group-mediated histone H2A monoubiquitination in epigenome regulation and nuclear processes.Nat Commun. 2020 Nov 23;11(1):5947. doi: 10.1038/s41467-020-19722-9. Nat Commun. 2020. PMID: 33230107 Free PMC article. Review.
Cited by
-
Using human disease mutations to understand de novo DNA methyltransferase function.Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review.
-
The N-terminal region of DNMT3A engages the nucleosome surface to aid chromatin recruitment.EMBO Rep. 2024 Dec;25(12):5743-5779. doi: 10.1038/s44319-024-00306-3. Epub 2024 Nov 11. EMBO Rep. 2024. PMID: 39528729 Free PMC article.
References
-
- Feinberg A. P., Cui H., Ohlsson R., DNA methylation and genomic imprinting: Insights from cancer into epigenetic mechanisms. Semin. Cancer Biol. 12, 389–398 (2002). - PubMed
MeSH terms
Substances
Grants and funding
- R44 HG011875/HG/NHGRI NIH HHS/United States
- F30 CA224971/CA/NCI NIH HHS/United States
- R35 GM138181/GM/NIGMS NIH HHS/United States
- R44 CA212733/CA/NCI NIH HHS/United States
- P01 CA098101/CA/NCI NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- R01 CA266978/CA/NCI NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
- R01 AA026297/AA/NIAAA NIH HHS/United States
- R01 DE031873/DE/NIDCR NIH HHS/United States
- R44 DE029633/DE/NIDCR NIH HHS/United States
- R44 GM119893/GM/NIGMS NIH HHS/United States
- S10 OD020056/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources

