Nuclear ATP-citrate lyase regulates chromatin-dependent activation and maintenance of the myofibroblast gene program
- PMID: 39196175
- PMCID: PMC11358007
- DOI: 10.1038/s44161-024-00502-3
Nuclear ATP-citrate lyase regulates chromatin-dependent activation and maintenance of the myofibroblast gene program
Abstract
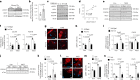
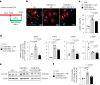
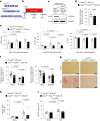
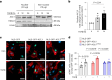
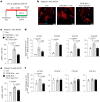
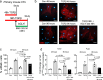
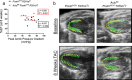
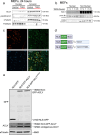
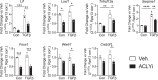
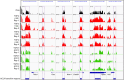
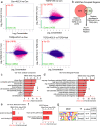
Differentiation of cardiac fibroblasts to myofibroblasts is necessary for matrix remodeling and fibrosis in heart failure. We previously reported that mitochondrial calcium signaling drives α-ketoglutarate-dependent histone demethylation, promoting myofibroblast formation. Here we investigate the role of ATP-citrate lyase (ACLY), a key enzyme for acetyl-CoA biosynthesis, in histone acetylation regulating myofibroblast fate and persistence in cardiac fibrosis. We show that inactivation of ACLY prevents myofibroblast differentiation and reverses myofibroblasts towards quiescence. Genetic deletion of Acly in post-activated myofibroblasts prevents fibrosis and preserves cardiac function in pressure-overload heart failure. TGFβ stimulation enhances ACLY nuclear localization and ACLY-SMAD2/3 interaction, and increases H3K27ac at fibrotic gene loci. Pharmacological inhibition of ACLY or forced nuclear expression of a dominant-negative ACLY mutant prevents myofibroblast formation and H3K27ac. Our data indicate that nuclear ACLY activity is necessary for myofibroblast differentiation and persistence by maintaining histone acetylation at TGFβ-induced myofibroblast genes. These findings provide targets to prevent and reverse pathological fibrosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Mammalian SIRT6 Represses Invasive Cancer Cell Phenotypes through ATP Citrate Lyase (ACLY)-Dependent Histone Acetylation.Genes (Basel). 2021 Sep 21;12(9):1460. doi: 10.3390/genes12091460. Genes (Basel). 2021. PMID: 34573442 Free PMC article.
-
Modulation of matrix metabolism by ATP-citrate lyase in articular chondrocytes.J Biol Chem. 2018 Aug 3;293(31):12259-12270. doi: 10.1074/jbc.RA118.002261. Epub 2018 Jun 21. J Biol Chem. 2018. PMID: 29929979 Free PMC article.
-
Fibroblast-specific TGF-β-Smad2/3 signaling underlies cardiac fibrosis.J Clin Invest. 2017 Oct 2;127(10):3770-3783. doi: 10.1172/JCI94753. Epub 2017 Sep 11. J Clin Invest. 2017. PMID: 28891814 Free PMC article.
-
Exploring the Role of ATP-Citrate Lyase in the Immune System.Front Immunol. 2021 Feb 18;12:632526. doi: 10.3389/fimmu.2021.632526. eCollection 2021. Front Immunol. 2021. PMID: 33679780 Free PMC article. Review.
-
ATP-citrate lyase: a key player in cancer metabolism.Cancer Res. 2012 Aug 1;72(15):3709-14. doi: 10.1158/0008-5472.CAN-11-4112. Epub 2012 Jul 10. Cancer Res. 2012. PMID: 22787121 Review.
Cited by
-
Metabolic control of collagen synthesis.Matrix Biol. 2024 Nov;133:43-56. doi: 10.1016/j.matbio.2024.07.003. Epub 2024 Jul 29. Matrix Biol. 2024. PMID: 39084474 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
