Insights into the regulatory role of epigenetics in moyamoya disease: Current advances and future prospectives
- PMID: 39188306
- PMCID: PMC11345382
- DOI: 10.1016/j.omtn.2024.102281
Insights into the regulatory role of epigenetics in moyamoya disease: Current advances and future prospectives
Abstract
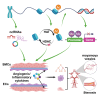
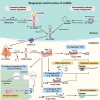
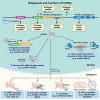
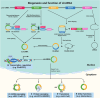
Moyamoya disease (MMD) is a progressive steno-occlusive cerebrovascular disorder that predominantly affecting East Asian populations. The intricate interplay of distinct and overlapping mechanisms, including genetic associations such as the RNF213-p.R4810K variant, contributes to the steno-occlusive lesions and moyamoya vessels. However, genetic mutations alone do not fully elucidate the occurrence of MMD, suggesting a potential role for epigenetic factors. Accruing evidence has unveiled the regulatory role of epigenetic markers, including DNA methylation, histone modifications, and non-coding RNAs (ncRNAs), in regulating pivotal cellular and molecular processes implicated in the pathogenesis of MMD by modulating endothelial cells and smooth muscle cells. The profile of these epigenetic markers in cerebral vasculatures and circulation has been determined to identify potential diagnostic biomarkers and therapeutic targets. Furthermore, in vitro studies have demonstrated the multifaceted effects of modulating specific epigenetic markers on MMD pathogenesis. These findings hold great potential for the discovery of novel therapeutic targets, translational studies, and clinical applications. In this review, we comprehensively summarize the current understanding of epigenetic mechanisms, including DNA methylation, histone modifications, and ncRNAs, in the context of MMD. Furthermore, we discuss the potential challenges and opportunities that lie ahead in this rapidly evolving field.
Keywords: DNA methylation; MT: Novel therapeutic targets and biomarker development Special Issue; epigenetics; histone modification; moyamoya disease; non-coding RNA.
© 2024 The Authors.
Conflict of interest statement
The authors declare no conflict of interests.
Figures
Similar articles
-
Importance of RNF213 polymorphism on clinical features and long-term outcome in moyamoya disease.J Neurosurg. 2016 May;124(5):1221-7. doi: 10.3171/2015.4.JNS142900. Epub 2015 Oct 2. J Neurosurg. 2016. PMID: 26430847
-
Rare variants of RNF213 and moyamoya/non-moyamoya intracranial artery stenosis/occlusion disease risk: a meta-analysis and systematic review.Environ Health Prev Med. 2017 Nov 2;22(1):75. doi: 10.1186/s12199-017-0680-1. Environ Health Prev Med. 2017. PMID: 29165161 Free PMC article. Review.
-
RNF213 as the major susceptibility gene for Chinese patients with moyamoya disease and its clinical relevance.J Neurosurg. 2017 Apr;126(4):1106-1113. doi: 10.3171/2016.2.JNS152173. Epub 2016 Apr 29. J Neurosurg. 2017. PMID: 27128593
-
Molecular analysis of RNF213 gene for moyamoya disease in the Chinese Han population.PLoS One. 2012;7(10):e48179. doi: 10.1371/journal.pone.0048179. Epub 2012 Oct 23. PLoS One. 2012. PMID: 23110205 Free PMC article.
-
Moyamoya Disease and Spectrums of RNF213 Vasculopathy.Transl Stroke Res. 2020 Aug;11(4):580-589. doi: 10.1007/s12975-019-00743-6. Epub 2019 Oct 24. Transl Stroke Res. 2020. PMID: 31650369 Review.
References
-
- Kuroda S., Houkin K. Moyamoya disease: current concepts and future perspectives. Lancet Neurol. 2008;7:1056–1066. - PubMed
-
- Guo D.C., Papke C.L., Tran-Fadulu V., Regalado E.S., Avidan N., Johnson R.J., Kim D.H., Pannu H., Willing M.C., Sparks E., et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am. J. Hum. Genet. 2009;84:617–627. - PMC - PubMed
-
- Chen T., Wei W., Yu J., Xu S., Zhang J., Li X., Chen J. The Progression of Pathophysiology of Moyamoya Disease. Neurosurgery. 2023;93:502–509. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

