This is a preprint.
Protein Arginine Methylation of the Translation Initiation Factor eIF1A Increases Usage of a Near-cognate Start Codon
- PMID: 39185183
- PMCID: PMC11343201
- DOI: 10.1101/2024.08.16.608280
Protein Arginine Methylation of the Translation Initiation Factor eIF1A Increases Usage of a Near-cognate Start Codon
Abstract
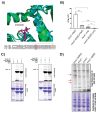
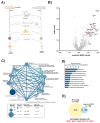
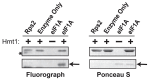
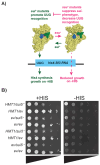
Protein arginine methylation has emerged as a key post-translational modification responsible for many facets of eukaryotic gene expression. To better understand the extent of this modification in cellular pathways, we carried out bioorthogonal methylation profiling in Saccharomyces cerevisiae to comprehensively identify the in vivo substrates of the major yeast protein arginine methyltransferase Hmt1. Gene ontology analysis of candidate substrates revealed an enrichment of proteins involved in the process of translation. We verified one such factor, eIF1A, by in vitro methylation. Three sites on eIF1A were found to be responsible for its methylation: R13, R14, and R62, with varied capacity by which each site contributed to the overall methylation capacity in vitro. To determine the role of methylation in eIF1A function, we used a battery of arginine-to-alanine substitution mutants to evaluate translation fidelity in these mutants. Our data show that substitution mutants at R13 and R14 in the N-terminal tail improved the fidelity of start codon recognition in an initiation fidelity assay. Overall, our data suggest that Hmt1-mediated methylation of eIF1A fine-tunes the fidelity of start codon recognition for proper translation initiation.
Figures


Similar articles
-
Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection.J Mol Biol. 2006 Feb 24;356(3):724-37. doi: 10.1016/j.jmb.2005.11.083. Epub 2005 Dec 15. J Mol Biol. 2006. PMID: 16380131
-
Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex.J Biol Chem. 2013 Feb 22;288(8):5316-29. doi: 10.1074/jbc.M112.440693. Epub 2013 Jan 4. J Biol Chem. 2013. PMID: 23293029 Free PMC article.
-
eIF1A residues implicated in cancer stabilize translation preinitiation complexes and favor suboptimal initiation sites in yeast.Elife. 2017 Dec 5;6:e31250. doi: 10.7554/eLife.31250. Elife. 2017. PMID: 29206102 Free PMC article.
-
Principles of start codon recognition in eukaryotic translation initiation.Nucleic Acids Res. 2016 Sep 30;44(17):8425-32. doi: 10.1093/nar/gkw534. Epub 2016 Jun 8. Nucleic Acids Res. 2016. PMID: 27280974 Free PMC article.
-
Should I stay or should I go? Eukaryotic translation initiation factors 1 and 1A control start codon recognition.J Biol Chem. 2008 Oct 10;283(41):27345-27349. doi: 10.1074/jbc.R800031200. Epub 2008 Jun 30. J Biol Chem. 2008. PMID: 18593708 Free PMC article. Review.
References
-
- Gary J. D., Lin W.-J., Yang M. C., Herschman H. R., and Clarke S. (1996) The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. Journal of Biological Chemistry 271, 12585–12594 - PubMed
-
- Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., and Herschman H. R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. Journal of Biological Chemistry 275, 7723–7730 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
