Spatial multi-omics: deciphering technological landscape of integration of multi-omics and its applications
- PMID: 39182134
- PMCID: PMC11344930
- DOI: 10.1186/s13045-024-01596-9
Spatial multi-omics: deciphering technological landscape of integration of multi-omics and its applications
Abstract
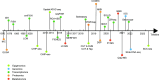
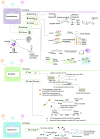
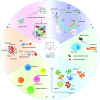
The emergence of spatial multi-omics has helped address the limitations of single-cell sequencing, which often leads to the loss of spatial context among cell populations. Integrated analysis of the genome, transcriptome, proteome, metabolome, and epigenome has enhanced our understanding of cell biology and the molecular basis of human diseases. Moreover, this approach offers profound insights into the interactions between intracellular and intercellular molecular mechanisms involved in the development, physiology, and pathogenesis of human diseases. In this comprehensive review, we examine current advancements in multi-omics technologies, focusing on their evolution and refinement over the past decade, including improvements in throughput and resolution, modality integration, and accuracy. We also discuss the pivotal contributions of spatial multi-omics in revealing spatial heterogeneity, constructing detailed spatial atlases, deciphering spatial crosstalk in tumor immunology, and advancing translational research and cancer therapy through precise spatial mapping.
Keywords: Crosstalk; Heterogeneity; Lineage tracking; New therapy; Reproduction; Spatial multi-omics; Spatial-specific atlas.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
The technological landscape and applications of single-cell multi-omics.Nat Rev Mol Cell Biol. 2023 Oct;24(10):695-713. doi: 10.1038/s41580-023-00615-w. Epub 2023 Jun 6. Nat Rev Mol Cell Biol. 2023. PMID: 37280296 Free PMC article. Review.
-
The burgeoning spatial multi-omics in human gastrointestinal cancers.PeerJ. 2024 Sep 13;12:e17860. doi: 10.7717/peerj.17860. eCollection 2024. PeerJ. 2024. PMID: 39285924 Free PMC article. Review.
-
Methods and applications for single-cell and spatial multi-omics.Nat Rev Genet. 2023 Aug;24(8):494-515. doi: 10.1038/s41576-023-00580-2. Epub 2023 Mar 2. Nat Rev Genet. 2023. PMID: 36864178 Free PMC article. Review.
-
Multi-omics based artificial intelligence for cancer research.Adv Cancer Res. 2024;163:303-356. doi: 10.1016/bs.acr.2024.06.005. Epub 2024 Jul 9. Adv Cancer Res. 2024. PMID: 39271266 Review.
-
Spatial multi-omics: novel tools to study the complexity of cardiovascular diseases.Genome Med. 2024 Jan 18;16(1):14. doi: 10.1186/s13073-024-01282-y. Genome Med. 2024. PMID: 38238823 Free PMC article. Review.
Cited by
-
Spatial immunogenomic patterns associated with lymph node metastasis in lung adenocarcinoma.Exp Hematol Oncol. 2024 Oct 28;13(1):106. doi: 10.1186/s40164-024-00574-8. Exp Hematol Oncol. 2024. PMID: 39468696 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

