Continuous exposure to doxorubicin induces stem cell-like characteristics and plasticity in MDA-MB-231 breast cancer cells identified with the SORE6 reporter
- PMID: 39180549
- PMCID: PMC11438702
- DOI: 10.1007/s00280-024-04701-4
Continuous exposure to doxorubicin induces stem cell-like characteristics and plasticity in MDA-MB-231 breast cancer cells identified with the SORE6 reporter
Abstract
Purpose: Cancer stem cells (CSCs) account for recurrence and resistance to breast cancer drugs, rendering them a cause of mortality and therapeutic failure. In this study, we examined the effects of exposure to low concentrations of doxorubicin (Dox) on CSCs and non-CSCs from TNBC.
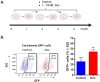
Methods: The effects of Dox were studied using the SORE6 reporter system. We examined the enrichment of the CSCs population, as well as the proliferation, and death of the reporter-positive fraction (GFP + cells) by flow cytometry. The resistant and stemness phenotypes were analyzed by viability and mammosphere formation assay, respectively. We identified differentially expressed and coregulated genes by RNA-seq analysis, and the correlation between gene expression and clinical outcome was evaluated by Kaplan-Mayer analysis using public databases.
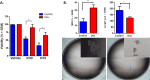
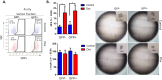
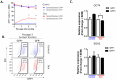
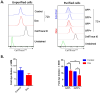
Results: In MDAMB231 and Hs578t cells, we identified enriched subsets in the CSCs population after continuous exposure to low concentrations of Dox. Cells from these enriched cultures showed resistance to toxic concentrations of Dox and increased efficiency of mammosphere formation. In purified GFP + or GFP- cells, Dox increased the mammosphere-forming efficiency, promoted phenotypic switches in non-CSCs populations to a CSC-like state, reduced proliferation, and induced differential gene expression. We identified several biological processes and molecular functions that partially explain the development of doxorubicin-resistant cells and cellular plasticity. Among the genes that were regulated by Dox exposure, the expression of ITGB1, SNAI1, NOTCH4, STAT5B, RAPGEF3, LAMA2, and GNAI1 was significantly associated with poor survival, the stemness phenotype, and chemoresistance.
Conclusion: The generation of chemoresistant cells that have characteristics of CSCs, after exposure to low concentrations of Dox, involves the differential expression of genes that have a clinical impact.
Keywords: Breast cancer; CSC-like; Doxorubicin; Drug-resistance; Plasticity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Stat3/Oct-4/c-Myc signal circuit for regulating stemness-mediated doxorubicin resistance of triple-negative breast cancer cells and inhibitory effects of WP1066.Int J Oncol. 2018 Jul;53(1):339-348. doi: 10.3892/ijo.2018.4399. Epub 2018 May 8. Int J Oncol. 2018. PMID: 29750424
-
Standard chemotherapy impacts on in vitro cellular heterogeneity in spheroids enriched with cancer stem cells (CSCs) derived from triple-negative breast cancer cell line.Biochem Biophys Res Commun. 2024 Nov 19;734:150765. doi: 10.1016/j.bbrc.2024.150765. Epub 2024 Sep 30. Biochem Biophys Res Commun. 2024. PMID: 39357337
-
Doxorubicin-Resistant TNBC Cells Exhibit Rapid Growth with Cancer Stem Cell-like Properties and EMT Phenotype, Which Can Be Transferred to Parental Cells through Autocrine Signaling.Int J Mol Sci. 2021 Nov 18;22(22):12438. doi: 10.3390/ijms222212438. Int J Mol Sci. 2021. PMID: 34830320 Free PMC article.
-
A live single-cell reporter system reveals drug-induced plasticity of a cancer stem cell-like population in cholangiocarcinoma.Sci Rep. 2024 Sep 30;14(1):22619. doi: 10.1038/s41598-024-73581-8. Sci Rep. 2024. PMID: 39349745 Free PMC article.
-
Regulation of stem cells-related signaling pathways in response to doxorubicin treatment in Hs578T triple-negative breast cancer cells.Mol Cell Biochem. 2015 Nov;409(1-2):163-76. doi: 10.1007/s11010-015-2522-z. Epub 2015 Jul 18. Mol Cell Biochem. 2015. PMID: 26187676
References
-
- Vučković L, Klisic A, Raonić J, Vućinić J (2021) Comparative study of immunohistochemical determination of breast cancer molecular subtypes on core biopsy and surgical specimens. Eur Rev Med Pharmacol Sci 25:3990–3996. 10.26355/eurrev_202106_26039 - PubMed
-
- Hammerl D, Smid M, Timmermans AM, Sleijfer S, Martens JWM, Debets R (2018) Breast cancer genomics and immuno-oncological markers to guide immune therapies. Semin Cancer Biol 52:178–188. 10.1016/j.semcancer.2017.11.003 - PubMed
-
- Waks AG, Winer EP (2019) Breast cancer treatment: a review. JAMA 321:288–300. 10.1001/JAMA.2018.19323 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

