Atomic resolution map of the solvent interactions driving SOD1 unfolding in CAPRIN1 condensates
- PMID: 39172789
- PMCID: PMC11363255
- DOI: 10.1073/pnas.2408554121
Atomic resolution map of the solvent interactions driving SOD1 unfolding in CAPRIN1 condensates
Abstract
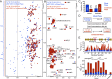
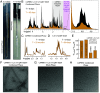
Biomolecules can be sequestered into membrane-less compartments, referred to as biomolecular condensates. Experimental and computational methods have helped define the physical-chemical properties of condensates. Less is known about how the high macromolecule concentrations in condensed phases contribute "solvent" interactions that can remodel the free-energy landscape of other condensate-resident proteins, altering thermally accessible conformations and, in turn, modulating function. Here, we use solution NMR spectroscopy to obtain atomic resolution insights into the interactions between the immature form of superoxide dismutase 1 (SOD1), which can mislocalize and aggregate in stress granules, and the RNA-binding protein CAPRIN1, a component of stress granules. NMR studies of CAPRIN1:SOD1 interactions, focused on both unfolded and folded SOD1 states in mixed phase and demixed CAPRIN1-based condensates, establish that CAPRIN1 shifts the SOD1 folding equilibrium toward the unfolded state through preferential interactions with the unfolded ensemble, with little change to the structure of the folded conformation. Key contacts between CAPRIN1 and the H80-H120 region of unfolded SOD1 are identified, as well as SOD1 interaction sites near both the arginine-rich and aromatic-rich regions of CAPRIN1. Unfolding of immature SOD1 in the CAPRIN1 condensed phase is shown to be coupled to aggregation, while a more stable zinc-bound, dimeric form of SOD1 is less susceptible to unfolding when solvated by CAPRIN1. Our work underscores the impact of the condensate solvent environment on the conformational states of resident proteins and supports the hypothesis that ALS mutations that decrease metal binding or dimerization function as drivers of aggregation in condensates.
Keywords: client/scaffold; folding-unfolding equilibria; methyl-TROSY NMR; phase separation; protein stability.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



Similar articles
-
Sequestration of Proteins in Stress Granules Relies on the In-Cell but Not the In Vitro Folding Stability.J Am Chem Soc. 2021 Dec 1;143(47):19909-19918. doi: 10.1021/jacs.1c09589. Epub 2021 Nov 17. J Am Chem Soc. 2021. PMID: 34788540
-
A delayed decoupling methyl-TROSY pulse sequence for atomic resolution studies of folded proteins and RNAs in condensates.J Magn Reson. 2024 May;362:107667. doi: 10.1016/j.jmr.2024.107667. Epub 2024 Mar 28. J Magn Reson. 2024. PMID: 38626504
-
Interaction hot spots for phase separation revealed by NMR studies of a CAPRIN1 condensed phase.Proc Natl Acad Sci U S A. 2021 Jun 8;118(23):e2104897118. doi: 10.1073/pnas.2104897118. Proc Natl Acad Sci U S A. 2021. PMID: 34074792 Free PMC article.
-
What are the distinguishing features and size requirements of biomolecular condensates and their implications for RNA-containing condensates?RNA. 2022 Jan;28(1):36-47. doi: 10.1261/rna.079026.121. Epub 2021 Nov 12. RNA. 2022. PMID: 34772786 Free PMC article. Review.
-
Using quantitative reconstitution to investigate multicomponent condensates.RNA. 2022 Jan;28(1):27-35. doi: 10.1261/rna.079008.121. Epub 2021 Nov 12. RNA. 2022. PMID: 34772789 Free PMC article. Review.
Cited by
-
A coarse-grained model for disordered and multi-domain proteins.Protein Sci. 2024 Nov;33(11):e5172. doi: 10.1002/pro.5172. Protein Sci. 2024. PMID: 39412378 Free PMC article.
References
-
- Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). - PubMed
MeSH terms
Substances
Grants and funding
- FND-503573/Canadian Government | Canadian Institutes of Health Research (CIHR)
- 2015-04347/Canadian Government | Natural Sciences and Engineering Research Council of Canada (NSERC)
- FDN-148375/Canadian Government | Canadian Institutes of Health Research (CIHR)
- PJT-190060/Canadian Government | Canadian Institutes of Health Research (CIHR)
LinkOut - more resources
Full Text Sources
Miscellaneous

