Genetic evidence for a regulated cysteine protease catalytic triad in LegA7, a Legionella pneumophila protein that impinges on a stress response pathway
- PMID: 39166849
- PMCID: PMC11423584
- DOI: 10.1128/msphere.00222-24
Genetic evidence for a regulated cysteine protease catalytic triad in LegA7, a Legionella pneumophila protein that impinges on a stress response pathway
Abstract
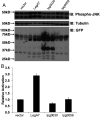
Legionella pneumophila grows within membrane-bound vacuoles in phylogenetically diverse hosts. Intracellular growth requires the function of the Icm/Dot type-IVb secretion system, which translocates more than 300 proteins into host cells. A screen was performed to identify L. pneumophila proteins that stimulate mitogen-activated protein kinase (MAPK) activation, using Icm/Dot translocated proteins ectopically expressed in mammalian cells. In parallel, a second screen was performed to identify L. pneumophila proteins expressed in yeast that cause growth inhibition in MAPK pathway-stimulatory high-osmolarity medium. LegA7 was shared in both screens, a protein predicted to be a member of the bacterial cysteine protease family that has five carboxyl-terminal ankyrin repeats. Three conserved residues in the predicted catalytic triad of LegA7 were mutated. These mutations abolished the ability of LegA7 to inhibit yeast growth. To identify other residues important for LegA7 function, a generalizable selection strategy in yeast was devised to isolate mutants that have lost function and no longer cause growth inhibition on a high-osmolarity medium. Mutations were isolated in the two carboxyl-terminal ankyrin repeats, as well as an inter-domain region located between the cysteine protease domain and the ankyrin repeats. These mutations were predicted by AlphaFold modeling to localize to the face opposite from the catalytic site, arguing that they interfere with the positive regulation of the catalytic activity. Based on our data, we present a model in which LegA7 harbors a cysteine protease domain with an inter-domain and two carboxyl-terminal ankyrin repeat regions that modulate the function of the catalytic domain.
Importance: Legionella pneumophila grows in a membrane-bound compartment in macrophages during disease. Construction of the compartment requires a dedicated secretion system that translocates virulence proteins into host cells. One of these proteins, LegA7, is shown to activate a stress response pathway in host cells called the mitogen-activated protein kinase (MAPK) pathway. The effects on the mammalian MAPK pathway were reconstructed in yeast, allowing the development of a strategy to identify the role of individual domains of LegA7. A domain similar to cysteine proteases is demonstrated to be critical for impinging on the MAPK pathway, and the catalytic activity of this domain is required for targeting this path. In addition, a conserved series of repeats, called ankyrin repeats, controls this activity. Data are provided that argue the interaction of the ankyrin repeats with unknown targets probably results in activation of the cysteine protease domain.
Keywords: HOG pathway; Legionella pneumophila; MAP kinases; Saccharomyces cerevisiae; ankyrin repeats; cysteine proteases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Update of
-
Genetic evidence for a regulated cysteine protease catalytic triad in LegA7, a Legionella pneumophila protein that impinges on a stress response pathway.bioRxiv [Preprint]. 2024 Jun 25:2024.03.17.585421. doi: 10.1101/2024.03.17.585421. bioRxiv. 2024. Update in: mSphere. 2024 Sep 25;9(9):e0022224. doi: 10.1128/msphere.00222-24. PMID: 38562771 Free PMC article. Updated. Preprint.
Similar articles
-
Genetic evidence for a regulated cysteine protease catalytic triad in LegA7, a Legionella pneumophila protein that impinges on a stress response pathway.bioRxiv [Preprint]. 2024 Jun 25:2024.03.17.585421. doi: 10.1101/2024.03.17.585421. bioRxiv. 2024. Update in: mSphere. 2024 Sep 25;9(9):e0022224. doi: 10.1128/msphere.00222-24. PMID: 38562771 Free PMC article. Updated. Preprint.
-
A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system.Mol Microbiol. 2005 May;56(4):918-33. doi: 10.1111/j.1365-2958.2005.04595.x. Mol Microbiol. 2005. PMID: 15853880
-
Interaction of the Ankyrin H Core Effector of Legionella with the Host LARP7 Component of the 7SK snRNP Complex.mBio. 2019 Aug 27;10(4):e01942-19. doi: 10.1128/mBio.01942-19. mBio. 2019. PMID: 31455655 Free PMC article.
-
Secreted phospholipases of the lung pathogen Legionella pneumophila.Int J Med Microbiol. 2018 Jan;308(1):168-175. doi: 10.1016/j.ijmm.2017.10.002. Epub 2017 Oct 28. Int J Med Microbiol. 2018. PMID: 29108710 Review.
-
Effector proteins translocated by Legionella pneumophila: strength in numbers.Trends Microbiol. 2007 Aug;15(8):372-80. doi: 10.1016/j.tim.2007.06.006. Epub 2007 Jul 13. Trends Microbiol. 2007. PMID: 17632005 Review.
References
MeSH terms
Substances
Grants and funding
- F32AI074193/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- T32GM008448/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- T32 GM008448/GM/NIGMS NIH HHS/United States
- F32-AI084202/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- F32 AI084202/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
