Modeling the roles of cohesotaxis, cell-intercalation, and tissue geometry in collective cell migration of Xenopus mesendoderm
- PMID: 39162010
- PMCID: PMC11360141
- DOI: 10.1242/bio.060615
Modeling the roles of cohesotaxis, cell-intercalation, and tissue geometry in collective cell migration of Xenopus mesendoderm
Abstract
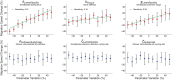
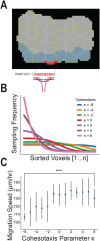
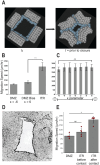
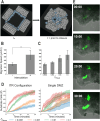
Collectively migrating Xenopus mesendoderm cells are arranged into leader and follower rows with distinct adhesive properties and protrusive behaviors. In vivo, leading row mesendoderm cells extend polarized protrusions and migrate along a fibronectin matrix assembled by blastocoel roof cells. Traction stresses generated at the leading row result in the pulling forward of attached follower row cells. Mesendoderm explants removed from embryos provide an experimentally tractable system for characterizing collective cell movements and behaviors, yet the cellular mechanisms responsible for this mode of migration remain elusive. We introduce a novel agent-based computational model of migrating mesendoderm in the Cellular-Potts computational framework to investigate the respective contributions of multiple parameters specific to the behaviors of leader and follower row cells. Sensitivity analyses identify cohesotaxis, tissue geometry, and cell intercalation as key parameters affecting the migration velocity of collectively migrating cells. The model predicts that cohesotaxis and tissue geometry in combination promote cooperative migration of leader cells resulting in increased migration velocity of the collective. Radial intercalation of cells towards the substrate is an additional mechanism contributing to an increase in migratory speed of the tissue. Model outcomes are validated experimentally using mesendoderm tissue explants.
Keywords: Agent-based model; CompuCell3D; Emergent property; Gastrulation; Mechanobiology; Tissue morphogenesis.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures


Update of
-
Modeling the roles of cohesotaxis, cell-intercalation, and tissue geometry in collective cell migration of Xenopus mesendoderm.bioRxiv [Preprint]. 2023 Oct 17:2023.10.16.562601. doi: 10.1101/2023.10.16.562601. bioRxiv. 2023. Update in: Biol Open. 2024 Aug 15;13(8):bio060615. doi: 10.1242/bio.060615. PMID: 37904937 Free PMC article. Updated. Preprint.
Similar articles
-
Modeling the roles of cohesotaxis, cell-intercalation, and tissue geometry in collective cell migration of Xenopus mesendoderm.bioRxiv [Preprint]. 2023 Oct 17:2023.10.16.562601. doi: 10.1101/2023.10.16.562601. bioRxiv. 2023. Update in: Biol Open. 2024 Aug 15;13(8):bio060615. doi: 10.1242/bio.060615. PMID: 37904937 Free PMC article. Updated. Preprint.
-
Mechanical and signaling roles for keratin intermediate filaments in the assembly and morphogenesis of Xenopus mesendoderm tissue at gastrulation.Development. 2017 Dec 1;144(23):4363-4376. doi: 10.1242/dev.155200. Epub 2017 Oct 5. Development. 2017. PMID: 28982683 Free PMC article.
-
Microsurgical Manipulations to Isolate Collectively Migrating Mesendoderm.Cold Spring Harb Protoc. 2022 Nov 1;2022(11):Pdb.prot097378. doi: 10.1101/pdb.prot097378. Cold Spring Harb Protoc. 2022. PMID: 35577524 Free PMC article.
-
Cell migration in the Xenopus gastrula.Wiley Interdiscip Rev Dev Biol. 2018 Nov;7(6):e325. doi: 10.1002/wdev.325. Epub 2018 Jun 26. Wiley Interdiscip Rev Dev Biol. 2018. PMID: 29944210 Review.
-
Mesoderm and endoderm internalization in the Xenopus gastrula.Curr Top Dev Biol. 2020;136:243-270. doi: 10.1016/bs.ctdb.2019.09.002. Epub 2019 Nov 5. Curr Top Dev Biol. 2020. PMID: 31959290 Review.
References
-
- Bornschlögl, T., Romero, S., Vestergaard, C. L., Joanny, J.-F., Van Nhieu, G. T. and Bassereau, P. (2013). Filopodial retraction force is generated by cortical actin dynamics and controlled by reversible tethering at the tip. Proc. Natl. Acad. Sci. USA 110, 18928-18933. 10.1073/pnas.1316572110 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

