Histone demethylase KDM2A recruits HCFC1 and E2F1 to orchestrate male germ cell meiotic entry and progression
- PMID: 39160277
- PMCID: PMC11448500
- DOI: 10.1038/s44318-024-00203-4
Histone demethylase KDM2A recruits HCFC1 and E2F1 to orchestrate male germ cell meiotic entry and progression
Abstract
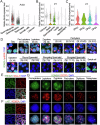
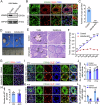
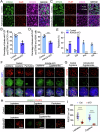
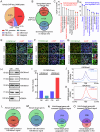
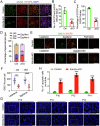
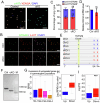
In mammals, the transition from mitosis to meiosis facilitates the successful production of gametes. However, the regulatory mechanisms that control meiotic initiation remain unclear, particularly in the context of complex histone modifications. Herein, we show that KDM2A, acting as a lysine demethylase targeting H3K36me3 in male germ cells, plays an essential role in modulating meiotic entry and progression. Conditional deletion of Kdm2a in mouse pre-meiotic germ cells results in complete male sterility, with spermatogenesis ultimately arrested at the zygotene stage of meiosis. KDM2A deficiency disrupts H3K36me2/3 deposition in c-KIT+ germ cells, characterized by a reduction in H3K36me2 but a dramatic increase in H3K36me3. Furthermore, KDM2A recruits the transcription factor E2F1 and its co-factor HCFC1 to the promoters of key genes required for meiosis entry and progression, such as Stra8, Meiosin, Spo11, and Sycp1. Collectively, our study unveils an essential role for KDM2A in mediating H3K36me2/3 deposition and controlling the programmed gene expression necessary for the transition from mitosis to meiosis during spermatogenesis.
Keywords: Fertility; Histone Modification; KDM2A; Male Germ Cells; Meiosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice.Proc Natl Acad Sci U S A. 2008 Sep 30;105(39):14976-80. doi: 10.1073/pnas.0807297105. Epub 2008 Sep 17. Proc Natl Acad Sci U S A. 2008. PMID: 18799751 Free PMC article.
-
KDM2A and KDM2B protect a subset of CpG islands from DNA methylation.J Genet Genomics. 2025 Jan;52(1):39-50. doi: 10.1016/j.jgg.2024.10.012. Epub 2024 Nov 8. J Genet Genomics. 2025. PMID: 39522683
-
KDM6A facilitates Xist upregulation at the onset of X inactivation.Biol Sex Differ. 2025 Jan 3;16(1):1. doi: 10.1186/s13293-024-00683-3. Biol Sex Differ. 2025. PMID: 39754175 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Context-Dependent and Locus-Specific Role of H3K36 Methylation in Transcriptional Regulation.J Mol Biol. 2025 Jan 1;437(1):168796. doi: 10.1016/j.jmb.2024.168796. Epub 2024 Sep 19. J Mol Biol. 2025. PMID: 39299382 Review.
References
-
- Adams IR, Davies OR (2023) Meiotic chromosome structure, the synaptonemal complex, and infertility. Annu Rev Genomics Hum Genet 24:35–61 - PubMed
-
- da Cruz I, Rodriguez-Casuriaga R, Santinaque FF, Farias J, Curti G, Capoano CA, Folle GA, Benavente R, Sotelo-Silveira JR, Geisinger A (2016) Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics 17:294 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

