CHC22 clathrin recruitment to the early secretory pathway requires two-site interaction with SNX5 and p115
- PMID: 39160272
- PMCID: PMC11445476
- DOI: 10.1038/s44318-024-00198-y
CHC22 clathrin recruitment to the early secretory pathway requires two-site interaction with SNX5 and p115
Abstract
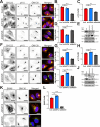
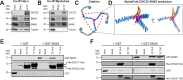
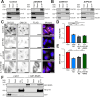
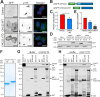
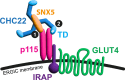
The two clathrin isoforms, CHC17 and CHC22, mediate separate intracellular transport routes. CHC17 performs endocytosis and housekeeping membrane traffic in all cells. CHC22, expressed most highly in skeletal muscle, shuttles the glucose transporter GLUT4 from the ERGIC (endoplasmic-reticulum-to-Golgi intermediate compartment) directly to an intracellular GLUT4 storage compartment (GSC), from where GLUT4 can be mobilized to the plasma membrane by insulin. Here, molecular determinants distinguishing CHC22 from CHC17 trafficking are defined. We show that the C-terminal trimerization domain of CHC22 interacts with SNX5, which also binds the ERGIC tether p115. SNX5, and the functionally redundant SNX6, are required for CHC22 localization independently of their participation in the endosomal ESCPE-1 complex. In tandem, an isoform-specific patch in the CHC22 N-terminal domain separately mediates binding to p115. This dual mode of clathrin recruitment, involving interactions at both N- and C-termini of the heavy chain, is required for CHC22 targeting to ERGIC membranes to mediate the Golgi-bypass route for GLUT4 trafficking. Interference with either interaction inhibits GLUT4 targeting to the GSC, defining a bipartite mechanism regulating a key pathway in human glucose metabolism.
Keywords: CHC22; Clathrin; Golgi bypass; Sorting nexin 5; p115.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
CHC22 clathrin mediates traffic from early secretory compartments for human GLUT4 pathway biogenesis.J Cell Biol. 2020 Jan 6;219(1):e201812135. doi: 10.1083/jcb.201812135. J Cell Biol. 2020. PMID: 31863584 Free PMC article.
-
Peer Play.2023 Jul 4. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 Jul 4. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30020595 Free Books & Documents.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
EMS Diabetic Protocols For Treat and Release.2023 Jul 17. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 Jul 17. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 32809447 Free Books & Documents.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
References
-
- Bogan JS (2012) Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem 81:507–532 - PubMed
-
- Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232 - PubMed
-
- Briant K, Redlingshöfer L, Brodsky FM (2020) Clathrin’s life beyond 40: connecting biochemistry with physiology and disease. Curr Opin Cell Biol 65:141–149 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

