Genome-wide sequencing identified extrachromosomal circular DNA as a transcription factor-binding motif of the senescence genes that govern replicative senescence in human mesenchymal stem cells
- PMID: 39157757
- PMCID: PMC11327076
- DOI: 10.3389/fncel.2024.1421342
Genome-wide sequencing identified extrachromosomal circular DNA as a transcription factor-binding motif of the senescence genes that govern replicative senescence in human mesenchymal stem cells
Abstract
Introduction: Mesenchymal stem cells (MSCs) have long been postulated as an important source cell in regenerative medicine. During subculture expansion, mesenchymal stem cell (MSC) senescence diminishes their multi-differentiation capabilities, leading to a loss of therapeutic potential. Up to date, the extrachromosomal circular DNAs (eccDNAs) have been demonstrated to be involved in senescence but the roles of eccDNAs during MSC.
Methods: Here we explored eccDNA profiles in human bone marrow MSCs (BM-MSCs). EccDNA and mRNA was purified and sequenced, followed by quantification and functional annotation. Moreover, we mapped our datasets with the downloading enhancer and transcription factor-regulated genes to explore the potential role of eccDNAs.
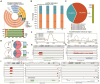
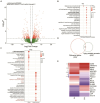
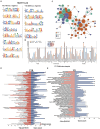
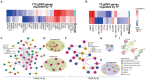
Results: Sequentially, gene annotation analysis revealed that the majority of eccDNA were mapped in the intron regions with limited BM-MSC enhancer overlaps. We discovered that these eccDNA motifs in senescent BMSCs acted as motifs for binding transcription factors (TFs) of senescence-related genes.
Discussion: These findings are highly significant for identifying biomarkers of senescence and therapeutic targets in mesenchymal stem cells (MSCs) for future clinical applications. The potential of eccDNA as a stable therapeutic target for senescence-related disorders warrants further investigation, particularly exploring chemically synthesized eccDNAs as transcription factor regulatory elements to reverse cellular senescence.
Keywords: biomarker; extrachromosomal circular DNAs; mesenchymal stem cells; senescence; transcription factors.
Copyright © 2024 Yang, Ji, Liao, Li, Wang, Lin, Wang and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Identification and characterization of extrachromosomal circular DNA in the silk gland of Bombyx mori.Insect Sci. 2023 Dec;30(6):1565-1578. doi: 10.1111/1744-7917.13191. Epub 2023 Apr 10. Insect Sci. 2023. PMID: 36826848
-
Extrachromosomal circular DNAs are common and functional in esophageal squamous cell carcinoma.Ann Transl Med. 2021 Sep;9(18):1464. doi: 10.21037/atm-21-4372. Ann Transl Med. 2021. PMID: 34734016 Free PMC article.
-
Identification and characterization of extrachromosomal circular DNA in large-artery atherosclerotic stroke.J Cell Mol Med. 2024 Apr;28(7):e18210. doi: 10.1111/jcmm.18210. J Cell Mol Med. 2024. PMID: 38506071 Free PMC article.
-
Extrachromosomal Circular DNAs: Origin, formation and emerging function in Cancer.Int J Biol Sci. 2021 Mar 2;17(4):1010-1025. doi: 10.7150/ijbs.54614. eCollection 2021. Int J Biol Sci. 2021. PMID: 33867825 Free PMC article. Review.
-
Extrachromosomal circular DNA: A neglected nucleic acid molecule in plants.Curr Opin Plant Biol. 2022 Oct;69:102263. doi: 10.1016/j.pbi.2022.102263. Epub 2022 Jul 21. Curr Opin Plant Biol. 2022. PMID: 35872391 Review.
References
-
- Anderson D. H., Radeke M. J., Gallo N. B., Chapin E. A., Johnson P. T., Curletti C. R., et al. . (2010). The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog. Retin. Eye Res. 29, 95–112. doi: 10.1016/j.preteyeres.2009.11.003, PMID: - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

