This is a preprint.
An asymmetric nautilus-like HflK/C assembly controls FtsH proteolysis of membrane proteins
- PMID: 39149393
- PMCID: PMC11326279
- DOI: 10.1101/2024.08.09.604662
An asymmetric nautilus-like HflK/C assembly controls FtsH proteolysis of membrane proteins
Abstract
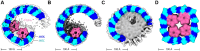
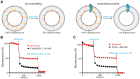
FtsH, a AAA protease, associates with HflK/C subunits to form a megadalton complex that spans the inner membrane and extends into the periplasm of E. coli. How this complex and homologous assemblies in eukaryotic organelles recruit, extract, and degrade membrane-embedded substrates is unclear. Following overproduction of protein components, recent cryo-EM structures reveal symmetric HflK/C cages surrounding FtsH in a manner proposed to inhibit degradation of membrane-embedded substrates. Here, we present structures of native complexes in which HflK/C instead forms an asymmetric nautilus-like assembly with an entryway for membrane-embedded substrates to reach and be engaged by FtsH. Consistent with this nautilus-like structure, proteomic assays suggest that HflK/C enhances FtsH degradation of certain membrane-embedded substrates. The membrane curvature in our FtsH•HflK/C complexes is opposite that of surrounding membrane regions, a property that correlates with lipid-scramblase activity and possibly with FtsH's function in the degradation of membrane-embedded proteins.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interest.
Figures



Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome.Bio Protoc. 2024 May 5;14(9):e4981. doi: 10.21769/BioProtoc.4981. eCollection 2024 May 5. Bio Protoc. 2024. PMID: 38737506 Free PMC article.
-
Dynamic Field Theory of Executive Function: Identifying Early Neurocognitive Markers.Monogr Soc Res Child Dev. 2024 Dec;89(3):7-109. doi: 10.1111/mono.12478. Monogr Soc Res Child Dev. 2024. PMID: 39628288 Free PMC article.
-
The phytoplasma SAP54 effector acts as a molecular matchmaker for leafhopper vectors by targeting plant MADS-box factor SVP.Elife. 2025 Jan 7;13:RP98992. doi: 10.7554/eLife.98992. Elife. 2025. PMID: 39763298 Free PMC article.
-
Exploring conceptual and theoretical frameworks for nurse practitioner education: a scoping review protocol.JBI Database System Rev Implement Rep. 2015 Oct;13(10):146-55. doi: 10.11124/jbisrir-2015-2150. JBI Database System Rev Implement Rep. 2015. PMID: 26571290
References
-
- Arends J, Thomanek N, Kuhlmann K, Marcus K, Narberhaus F (2016) In vivo trapping of FtsH substrates by label-free quantitative proteomics. Proteomics 16: 3161–3172 - PubMed
-
- Bittner LM, Arends J, Narberhaus F (2017) When, how and why? Regulated proteolysis by the essential FtsH protease in Escherichia coli. Biol Chem 398: 625–635 - PubMed
-
- Browman DT, Hoegg MB, Robbins SM (2007) The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol 17: 394–402 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
