Multiscale simulations reveal the driving forces of p53C phase separation accelerated by oncogenic mutations
- PMID: 39148776
- PMCID: PMC11323318
- DOI: 10.1039/d4sc03645j
Multiscale simulations reveal the driving forces of p53C phase separation accelerated by oncogenic mutations
Abstract
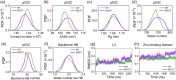
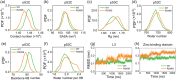
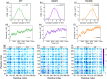
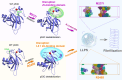
Liquid-Liquid phase separation (LLPS) of p53 to form liquid condensates has been implicated in cellular functions and dysfunctions. The p53 condensates may serve as amyloid fibril precursors to initiate p53 aggregation, which is associated with oncogenic gain-of-function and various human cancers. M237I and R249S mutations located in p53 core domain (p53C) have been detected respectively in glioblastomas and hepatocellular carcinoma. Interestingly, these p53C mutants can also undergo LLPS and liquid-to-solid phase transition, which are faster than wild type p53C. However, the underlying molecular basis governing the accelerated LLPS and liquid-to-solid transition of p53C remain poorly understood. Herein, we explore the M237I/R249S mutation-induced structural alterations and phase separation behavior of p53C by employing multiscale molecular dynamics simulations. All-atom simulations revealed conformational disruptions in the zinc-binding domain of the M237I mutant and in both loop3 and zinc-binding domain of the R249S mutant. The two mutations enhance hydrophobic exposure of those regions and attenuate intramolecular interactions, which may hasten the LLPS and aggregation of p53C. Martini 3 coarse-grained simulations demonstrated spontaneous phase separation of p53C and accelerated effects of M237I/R249S mutations on the phase separation of p53C. Importantly, we find that the regions with enhanced intermolecular interactions observed in coarse-grained simulations coincide with the disrupted regions with weakened intramolecular interactions observed in all-atom simulations, indicating that M237I/R249S mutation-induced local structural disruptions expedite the LLPS of p53C. This study unveils the molecular mechanisms underlying the two cancer-associated mutation-accelerated LLPS and aggregation of p53C, providing avenues for anticancer therapy by targeting the phase separation process.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




Similar articles
-
Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands.Chem Sci. 2021 Apr 26;12(21):7334-7349. doi: 10.1039/d1sc01739j. Chem Sci. 2021. PMID: 34163823 Free PMC article.
-
Oncogenic R248W mutation induced conformational perturbation of the p53 core domain and the structural protection by proteomimetic amyloid inhibitor ADH-6.Phys Chem Chem Phys. 2024 Jul 24;26(29):20068-20086. doi: 10.1039/d4cp02046d. Phys Chem Chem Phys. 2024. PMID: 39007865
-
Elucidating the Mechanisms of R248Q Mutation-Enhanced p53 Aggregation and Its Inhibition by Resveratrol.J Phys Chem B. 2023 Sep 14;127(36):7708-7720. doi: 10.1021/acs.jpcb.3c04700. Epub 2023 Sep 4. J Phys Chem B. 2023. PMID: 37665658
-
Liquid - liquid phase separation of tau: Driving forces, regulation, and biological implications.Neurobiol Dis. 2023 Jul;183:106167. doi: 10.1016/j.nbd.2023.106167. Epub 2023 May 23. Neurobiol Dis. 2023. PMID: 37230179 Review.
-
Challenges in studying the liquid-to-solid phase transitions of proteins using computer simulations.Curr Opin Chem Biol. 2023 Aug;75:102333. doi: 10.1016/j.cbpa.2023.102333. Epub 2023 May 31. Curr Opin Chem Biol. 2023. PMID: 37267850 Free PMC article. Review.
References
-
- Brangwynne C. P. Tompa P. Pappu R. V. Nat. Phys. 2015;11:899–904.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

