The role of protein post-translational modifications in prostate cancer
- PMID: 39148683
- PMCID: PMC11326433
- DOI: 10.7717/peerj.17768
The role of protein post-translational modifications in prostate cancer
Abstract
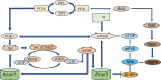
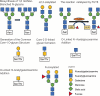
Involving addition of chemical groups or protein units to specific residues of the target protein, post-translational modifications (PTMs) alter the charge, hydrophobicity, and conformation of a protein, which in turn influences protein function, protein-protein interaction, and protein aggregation. These alterations, which include phosphorylation, glycosylation, ubiquitination, methylation, acetylation, lipidation, and lactylation, are significant biological events in the development of cancer, and play vital roles in numerous biological processes. The processes behind essential functions, the screening of clinical illness signs, and the identification of therapeutic targets all depend heavily on further research into the PTMs. This review outlines the influence of several PTM types on prostate cancer (PCa) diagnosis, therapy, and prognosis in an effort to shed fresh light on the molecular causes and progression of the disease.
Keywords: Diagnosis; Prognosis; Prostate cancer; Protein post-translational modification; Treatment.
©2024 Hao et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
Similar articles
-
The Role of Post-Translational Modifications on the Structure and Function of Tau Protein.J Mol Neurosci. 2022 Aug;72(8):1557-1571. doi: 10.1007/s12031-022-02002-0. Epub 2022 Mar 24. J Mol Neurosci. 2022. PMID: 35325356 Review.
-
Post-Translational Modifications That Drive Prostate Cancer Progression.Biomolecules. 2021 Feb 9;11(2):247. doi: 10.3390/biom11020247. Biomolecules. 2021. PMID: 33572160 Free PMC article. Review.
-
Advances in post-translational modifications of proteins and cancer immunotherapy.Front Immunol. 2023 Aug 22;14:1229397. doi: 10.3389/fimmu.2023.1229397. eCollection 2023. Front Immunol. 2023. PMID: 37675097 Free PMC article. Review.
-
A Text Mining and Machine Learning Protocol for Extracting Posttranslational Modifications of Proteins from PubMed: A Special Focus on Glycosylation, Acetylation, Methylation, Hydroxylation, and Ubiquitination.Methods Mol Biol. 2022;2496:179-202. doi: 10.1007/978-1-0716-2305-3_10. Methods Mol Biol. 2022. PMID: 35713865
-
Advances in Prediction of Posttranslational Modification Sites Known to Localize in Protein Supersecondary Structures.Methods Mol Biol. 2025;2870:117-151. doi: 10.1007/978-1-0716-4213-9_8. Methods Mol Biol. 2025. PMID: 39543034
References
-
- Baratchian M, Tiwari R, Khalighi S, Chakravarthy A, Yuan W, Berk M, Li J, Guerinot A, De Bono J, Makarov V, Chan TA, Silverman RH, Stark GR, Varadan V, De Carvalho DD, Chakraborty AA, Sharifi N. H3K9 methylation drives resistance to androgen receptor-antagonist therapy in prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 2022;119:e2114324119. doi: 10.1073/pnas.2114324119. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

