Characterization and transmission of plasmid-mediated multidrug resistance in foodborne Vibrio parahaemolyticus
- PMID: 39144225
- PMCID: PMC11322368
- DOI: 10.3389/fmicb.2024.1437660
Characterization and transmission of plasmid-mediated multidrug resistance in foodborne Vibrio parahaemolyticus
Abstract
Objectives: The purpose of this study was to determine the structural features and transferability of the multidrug-resistance (MDR) plasmid, and resistance phenotypes for the tested antimicrobials in foodborne Vibrio parahaemolyticus.
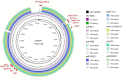
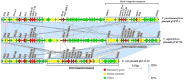
Methods: Plasmids were isolated from a V. parahaemolyticus strain of seafood origin, then sequenced using the Illumina NovaSeq 6000 and PacBio Sequel II sequencing platforms to obtain the complete genome data. Characterization of the MDR plasmid pVP52-1, including determination of antimicrobial resistance genes (ARGs), plasmid incompatibility groups, and transferability, was carried out.
Results: V. parahaemolyticus strain NJIFDCVp52 contained two circular chromosomes and two circular plasmids (pVP52-1 and pVP52-2). Plasmid typing indicated that pVP52-1 belonged to the incompatibility group IncA/C2 and the sequence type pST3. pVP52-1 carried 12 different ARGs, an IS110-composite transposon consisting of aac(6')-Ib-cr, qnrVC1, aac(6')-Ib, dfrA14, and the IS26-mphA-IS6100 unit flanked by inverted sequences of IS5075 and IS4321. pVP52-2 carried no ARGs. A plasmid elimination assay showed that only pVP52-1 and its ARGs were lost, the loss of resistance to several antimicrobials, causing a change from the ampicillin-ampicillin/sulbactam-cefazolin-cefoxitin-ceftazidime-cefotaxime-imipenem-trimethoprim/sulfamethoxazole resistance pattern to the ampicillin resistance pattern. In accordance, a conjugation transfer assay showed that only pVP52-1 and its ARGs were horizontally transferred, leading to increased antimicrobial resistance in Escherichia coli strain EC600, causing a change from the ampicillin-nalidixic acid resistance pattern to the ampicillin-ampicillin/sulbactam-cefazolin-cefoxitin-ceftazidime-cefotaxime-imipenem-nalidixic acid-chloramphenicol-tetracycline-trimethoprim/sulfamethoxazole-azithromycin resistance pattern. Further transferability experiments revealed that pVP52-1 could be transferred to other enterobacterial strains of E. coli and Salmonella.
Discussion: This study emphasizes the urgent need for continued surveillance of resistance plasmids and changes in antimicrobial resistance profiles among the V. parahaemolyticus population.
Keywords: Vibrio parahaemolyticus; antimicrobial resistance genes; horizontal transfer; multidrug resistance; plasmid; whole genome sequencing.
Copyright © 2024 Zhou, Lu, Liu, Bie, Cui, Wang, Sun and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Whole genome sequencing and antimicrobial resistance analysis of Vibrio parahaemolyticus Vp2015094 carrying an antimicrobial-resistant plasmid.J Glob Antimicrob Resist. 2022 Sep;30:47-49. doi: 10.1016/j.jgar.2022.05.025. Epub 2022 Jun 2. J Glob Antimicrob Resist. 2022. PMID: 35660661
-
Multidrug resistance and multivirulence plasmids in enterotoxigenic and hybrid Shiga toxin-producing/enterotoxigenic Escherichia coli isolated from diarrheic pigs in Switzerland.Vet J. 2019 Feb;244:60-68. doi: 10.1016/j.tvjl.2018.12.015. Epub 2018 Dec 12. Vet J. 2019. PMID: 30825896
-
Genomic Analysis of an Escherichia coli Sequence Type 167 Isolate Harboring a Multidrug-Resistant Conjugative Plasmid, Suggesting the Potential Transmission of the Type Strains from Animals to Humans.Infect Drug Resist. 2023 Aug 8;16:5077-5084. doi: 10.2147/IDR.S420635. eCollection 2023. Infect Drug Resist. 2023. PMID: 37576518 Free PMC article.
-
Prevalence, multidrug resistance, and biofilm formation of Vibrio parahaemolyticus isolated from fish mariculture environments in Cat Ba Island, Vietnam.Osong Public Health Res Perspect. 2024 Feb;15(1):56-67. doi: 10.24171/j.phrp.2023.0181. Epub 2024 Feb 19. Osong Public Health Res Perspect. 2024. PMID: 38481050 Free PMC article.
-
Incidence, genetic diversity, and antimicrobial resistance profiles of Vibrio parahaemolyticus in seafood in Bangkok and eastern Thailand.PeerJ. 2023 May 11;11:e15283. doi: 10.7717/peerj.15283. eCollection 2023. PeerJ. 2023. PMID: 37193031 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

