Major depletion of insulin sensitivity-associated taxa in the gut microbiome of persons living with HIV controlled by antiretroviral drugs
- PMID: 39138568
- PMCID: PMC11320835
- DOI: 10.1186/s12920-024-01978-5
Major depletion of insulin sensitivity-associated taxa in the gut microbiome of persons living with HIV controlled by antiretroviral drugs
Abstract
Background: Persons living with HIV (PWH) harbor an altered gut microbiome (higher abundance of Prevotella and lower abundance of Bacillota and Ruminococcus lineages) compared to non-infected individuals. Some of these alterations are linked to sexual preference and others to the HIV infection. The relationship between these lineages and metabolic alterations, often present in aging PWH, has been poorly investigated.
Methods: In this study, we compared fecal metagenomes of 25 antiretroviral-treatment (ART)-controlled PWH to three independent control groups of 25 non-infected matched individuals by means of univariate analyses and machine learning methods. Moreover, we used two external datasets to validate predictive models of PWH classification. Next, we searched for associations between clinical and biological metabolic parameters with taxonomic and functional microbiome profiles. Finally, we compare the gut microbiome in 7 PWH after a 17-week ART switch to raltegravir/maraviroc.
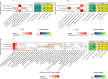
Results: Three major enterotypes (Prevotella, Bacteroides and Ruminococcaceae) were present in all groups. The first Prevotella enterotype was enriched in PWH, with several of characteristic lineages associated with poor metabolic profiles (low HDL and adiponectin, high insulin resistance (HOMA-IR)). Conversely butyrate-producing lineages were markedly depleted in PWH independently of sexual preference and were associated with a better metabolic profile (higher HDL and adiponectin and lower HOMA-IR). Accordingly with the worst metabolic status of PWH, butyrate production and amino-acid degradation modules were associated with high HDL and adiponectin and low HOMA-IR. Random Forest models trained to classify PWH vs. control on taxonomic abundances displayed high generalization performance on two external holdout datasets (ROC AUC of 80-82%). Finally, no significant alterations in microbiome composition were observed after switching to raltegravir/maraviroc.
Conclusion: High resolution metagenomic analyses revealed major differences in the gut microbiome of ART-controlled PWH when compared with three independent matched cohorts of controls. The observed marked insulin resistance could result both from enrichment in Prevotella lineages, and from the depletion in species producing butyrate and involved into amino-acid degradation, which depletion is linked with the HIV infection.
Keywords: HIV; Human gut microbiome; Machine learning; Metabolic status.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Gut resistome linked to sexual preference and HIV infection.BMC Microbiol. 2024 Jun 8;24(1):201. doi: 10.1186/s12866-024-03335-z. BMC Microbiol. 2024. PMID: 38851693 Free PMC article.
-
Sustained gut dysbiosis and intestinal inflammation show correlation with weight gain in person with chronic HIV infection on antiretroviral therapy.BMC Microbiol. 2024 Jul 24;24(1):274. doi: 10.1186/s12866-024-03431-0. BMC Microbiol. 2024. PMID: 39044127 Free PMC article.
-
Markers of metabolic health and gut microbiome diversity: findings from two population-based cohort studies.Diabetologia. 2021 Aug;64(8):1749-1759. doi: 10.1007/s00125-021-05464-w. Epub 2021 Jun 10. Diabetologia. 2021. PMID: 34110438 Free PMC article.
-
A Unique Gut Microbiome-Physical Function Axis Exists in Older People with HIV: An Exploratory Study.AIDS Res Hum Retroviruses. 2021 Jul;37(7):542-550. doi: 10.1089/AID.2020.0283. Epub 2021 Apr 30. AIDS Res Hum Retroviruses. 2021. PMID: 33787299 Free PMC article.
-
HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy.Gut Microbes. 2014 Jul 1;5(4):562-70. doi: 10.4161/gmic.32132. Epub 2014 Jul 31. Gut Microbes. 2014. PMID: 25078714 Review.
References
-
- Schouten J, Wit FW, Stolte IG, Kootstra NA, van der Valk M, Geerlings SE, et al. Cross-sectional comparison of the prevalence of age-associated comorbidities and their risk factors between HIV-infected and uninfected individuals: the AGEhIV cohort study. Clin Infect Dis. 2014;59:1787–97. 10.1093/cid/ciu701 - DOI - PubMed
-
- Bastard J-P, Couffignal C, Fellahi S, Bard J-M, Mentre F, Salmon D, et al. Diabetes and dyslipidaemia are associated with oxidative stress independently of inflammation in long-term antiretroviral-treated HIV-infected patients. Diabetes Metab. 2019;45:573–81. 10.1016/j.diabet.2019.02.008 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

