Involvement of tumor immune microenvironment metabolic reprogramming in colorectal cancer progression, immune escape, and response to immunotherapy
- PMID: 39119332
- PMCID: PMC11306065
- DOI: 10.3389/fimmu.2024.1353787
Involvement of tumor immune microenvironment metabolic reprogramming in colorectal cancer progression, immune escape, and response to immunotherapy
Abstract
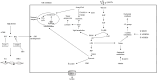
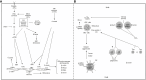
Metabolic reprogramming is a k`ey hallmark of tumors, developed in response to hypoxia and nutrient deficiency during tumor progression. In both cancer and immune cells, there is a metabolic shift from oxidative phosphorylation (OXPHOS) to aerobic glycolysis, also known as the Warburg effect, which then leads to lactate acidification, increased lipid synthesis, and glutaminolysis. This reprogramming facilitates tumor immune evasion and, within the tumor microenvironment (TME), cancer and immune cells collaborate to create a suppressive tumor immune microenvironment (TIME). The growing interest in the metabolic reprogramming of the TME, particularly its significance in colorectal cancer (CRC)-one of the most prevalent cancers-has prompted us to explore this topic. CRC exhibits abnormal glycolysis, glutaminolysis, and increased lipid synthesis. Acidosis in CRC cells hampers the activity of anti-tumor immune cells and inhibits the phagocytosis of tumor-associated macrophages (TAMs), while nutrient deficiency promotes the development of regulatory T cells (Tregs) and M2-like macrophages. In CRC cells, activation of G-protein coupled receptor 81 (GPR81) signaling leads to overexpression of programmed death-ligand 1 (PD-L1) and reduces the antigen presentation capability of dendritic cells. Moreover, the genetic and epigenetic cell phenotype, along with the microbiota, significantly influence CRC metabolic reprogramming. Activating RAS mutations and overexpression of epidermal growth factor receptor (EGFR) occur in approximately 50% and 80% of patients, respectively, stimulating glycolysis and increasing levels of hypoxia-inducible factor 1 alpha (HIF-1α) and MYC proteins. Certain bacteria produce short-chain fatty acids (SCFAs), which activate CD8+ cells and genes involved in antigen processing and presentation, while other mechanisms support pro-tumor activities. The use of immune checkpoint inhibitors (ICIs) in selected CRC patients has shown promise, and the combination of these with drugs that inhibit aerobic glycolysis is currently being intensively researched to enhance the efficacy of immunotherapy.
Keywords: colorectal cancer; immunotherapy; metabolic reprogramming; tumor immune escape; tumor immune microenvironment.
Copyright © 2024 Nicolini and Ferrari.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Metabolic Reprogramming of Colorectal Cancer Cells and the Microenvironment: Implication for Therapy.Int J Mol Sci. 2021 Jun 10;22(12):6262. doi: 10.3390/ijms22126262. Int J Mol Sci. 2021. PMID: 34200820 Free PMC article. Review.
-
Metabolic Factors Affecting Tumor Immunogenicity: What Is Happening at the Cellular Level?Int J Mol Sci. 2021 Feb 21;22(4):2142. doi: 10.3390/ijms22042142. Int J Mol Sci. 2021. PMID: 33670011 Free PMC article. Review.
-
T-cell immunoglobulin and ITIM domain, as a potential immune checkpoint target for immunotherapy of colorectal cancer.IUBMB Life. 2021 May;73(5):726-738. doi: 10.1002/iub.2461. Epub 2021 Mar 30. IUBMB Life. 2021. PMID: 33686787 Review.
-
The roles of epigallocatechin gallate in the tumor microenvironment, metabolic reprogramming, and immunotherapy.Front Immunol. 2024 Jan 29;15:1331641. doi: 10.3389/fimmu.2024.1331641. eCollection 2024. Front Immunol. 2024. PMID: 38348027 Free PMC article. Review.
-
Local TSH/TSHR signaling promotes CD8+ T cell exhaustion and immune evasion in colorectal carcinoma.Cancer Commun (Lond). 2024 Nov;44(11):1287-1310. doi: 10.1002/cac2.12605. Epub 2024 Sep 16. Cancer Commun (Lond). 2024. PMID: 39285586 Free PMC article.
Cited by
-
The Major Role of T Regulatory Cells in the Efficiency of Vaccination in General and Immunocompromised Populations: A Review.Vaccines (Basel). 2024 Aug 30;12(9):992. doi: 10.3390/vaccines12090992. Vaccines (Basel). 2024. PMID: 39340024 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

