siRNA-loaded folic acid-modified TPGS alleviate MASH via targeting ER stress sensor XBP1 and reprogramming macrophages
- PMID: 39113706
- PMCID: PMC11302883
- DOI: 10.7150/ijbs.96113
siRNA-loaded folic acid-modified TPGS alleviate MASH via targeting ER stress sensor XBP1 and reprogramming macrophages
Abstract
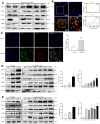
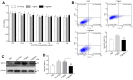
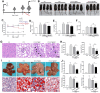
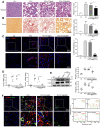
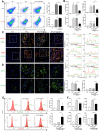
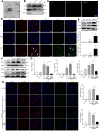
Macrophages show high plasticity and play a vital role in the progression of metabolic dysfunction-associated steatohepatitis (MASH). X-box binding protein 1 (XBP1), a key sensor of the unfolded protein response, can modulate macrophage-mediated pro-inflammatory responses in the pathogenesis of MASH. However, how XBP1 influences macrophage plasticity and promotes MASH progression remains unclear. Herein, we formulated an Xbp1 siRNA delivery system based on folic acid modified D-α-tocopheryl polyethylene glycol 1000 succinate nanoparticles (FT@XBP1) to explore the precise role of macrophage-specific Xbp1 deficiency in the progression of MASH. FT@XBP1 was specifically internalized into hepatic macrophages and subsequently inhibited the expression of spliced XBP1 both in vitro and in vivo. It promoted M1-phenotype macrophage repolarization to M2 macrophages, reduced the release of pro-inflammatory factors, and alleviated hepatic steatosis, liver injury, and fibrosis in mice with fat-, fructose- and cholesterol-rich diet-induced MASH. Mechanistically, FT@XBP1 promoted macrophage polarization toward the M2 phenotype and enhanced the release of exosomes that could inhibit the activation of hepatic stellate cells. A promising macrophage-targeted siRNA delivery system was revealed to pave a promising strategy in the treatment of MASH.
Keywords: D-α-Tocopheryl polyethylene glycol 1000 succinate; Endoplasmic reticulum stress; Macrophages; Metabolic dysfunction-associated steatohepatitis; X-box binding protein 1.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


Similar articles
-
The unfolded protein response regulates hepatic autophagy by sXBP1-mediated activation of TFEB.Autophagy. 2021 Aug;17(8):1841-1855. doi: 10.1080/15548627.2020.1788889. Epub 2020 Jul 15. Autophagy. 2021. PMID: 32597296 Free PMC article.
-
XBP1-mediated activation of the STING signalling pathway in macrophages contributes to liver fibrosis progression.JHEP Rep. 2022 Aug 18;4(11):100555. doi: 10.1016/j.jhepr.2022.100555. eCollection 2022 Nov. JHEP Rep. 2022. PMID: 36185574 Free PMC article.
-
Role of XBP1 in regulating the progression of non-alcoholic steatohepatitis.J Hepatol. 2022 Aug;77(2):312-325. doi: 10.1016/j.jhep.2022.02.031. Epub 2022 Mar 12. J Hepatol. 2022. PMID: 35292349
-
Macrophage ATG16L1 expression suppresses metabolic dysfunction-associated steatohepatitis progression by promoting lipophagy.Clin Mol Hepatol. 2024 Jul;30(3):515-538. doi: 10.3350/cmh.2024.0107. Epub 2024 May 10. Clin Mol Hepatol. 2024. PMID: 38726504 Free PMC article.
-
Hepatocyte X-box binding protein 1 deficiency increases liver injury in mice fed a high-fat/sugar diet.Am J Physiol Gastrointest Liver Physiol. 2015 Dec 15;309(12):G965-74. doi: 10.1152/ajpgi.00132.2015. Epub 2015 Oct 15. Am J Physiol Gastrointest Liver Physiol. 2015. PMID: 26472223 Free PMC article.
References
-
- Younossi ZM, Koenig AB, Abdelatif D. et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. - PubMed
-
- Romero-Gomez M, Zelber-Sagi S, Trenell M. Treatment of NAFLD with diet, physical activity and exercise. J Hepatol. 2017;67:829–46. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

