tRF-27 competitively Binds to G3BPs and Activates MTORC1 to Enhance HER2 Positive Breast Cancer Trastuzumab Tolerance
- PMID: 39113695
- PMCID: PMC11302882
- DOI: 10.7150/ijbs.87415
tRF-27 competitively Binds to G3BPs and Activates MTORC1 to Enhance HER2 Positive Breast Cancer Trastuzumab Tolerance
Abstract
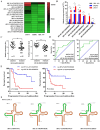
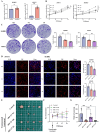
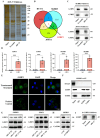
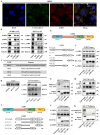
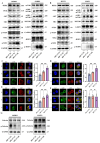
About 20% of breast cancer patients are positive for HER2. The efficacy of current treatments is limited by primary and secondary resistance to trastuzumab. tRNA-derived fragments (tRFs) have shown crucial regulatory roles in various cancers. This study aimed to evaluate the role of tRF-27 in regulating the resistance of HER2-positive breast cancer against trastuzumab. tRF-27 was highly expressed in trastuzumab-resistant cells, and its expression level could predict the resistance to trastuzumab. High expression of tRF-27 promoted the growth and proliferation of trastuzumab-exposed cells. RNA-pulldown assay and mass spectrometry were performed to identify Ras GTPase-activating protein-binding proteins 1 and 2 (G3BPs) (two proteins targeted by tRF-27); RNA-immunoprecipitation (RIP) to confirm their bindings; co-immunoprecipitation (co-IP) and RNA-pulldown assay to determine the binding domains between G3BPs and tRF-27.tRF-27 bound to the nuclear transport factor 2 like domain(NTF2 domain) of G3BPs through a specific sequence. tRF-27 relied on G3BPs and NTF2 domain to increase trastuzumab tolerance. tRF-27 competed with lysosomal associated membrane protein 1(LAMP1) for NTF2 domain, thereby inhibiting lysosomal localization of G3BPs and tuberous sclerosis complex (TSC). Overexpression of tRF-27 inhibited phosphorylation of TSCs and promoted the activation of mechanistic target of rapamycin complex 1(MTORC1) to enhance cell proliferation and entice the resistance of HER2-positive breast cancer against trastuzumab.
Keywords: G3BPs; HER2-positive breast cancer; MTORC1; Trastuzumab resistance; tRNA-derived fragment.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
tRNA-Derived Fragments as Novel Predictive Biomarkers for Trastuzumab-Resistant Breast Cancer.Cell Physiol Biochem. 2018;49(2):419-431. doi: 10.1159/000492977. Epub 2018 Aug 28. Cell Physiol Biochem. 2018. PMID: 30153663
-
Rasputin a decade on and more promiscuous than ever? A review of G3BPs.Biochim Biophys Acta Mol Cell Res. 2019 Mar;1866(3):360-370. doi: 10.1016/j.bbamcr.2018.09.001. Epub 2018 Sep 5. Biochim Biophys Acta Mol Cell Res. 2019. PMID: 30595162 Free PMC article. Review.
-
EPHA5 mediates trastuzumab resistance in HER2-positive breast cancers through regulating cancer stem cell-like properties.FASEB J. 2019 Apr;33(4):4851-4865. doi: 10.1096/fj.201701561RRRR. Epub 2019 Jan 8. FASEB J. 2019. PMID: 30620624
-
Targeted dual degradation of HER2 and EGFR obliterates oncogenic signaling, overcomes therapy resistance, and inhibits metastatic lesions in HER2-positive breast cancer models.Drug Resist Updat. 2024 May;74:101078. doi: 10.1016/j.drup.2024.101078. Epub 2024 Mar 13. Drug Resist Updat. 2024. PMID: 38503142
-
Role(s) of G3BPs in Human Pathogenesis.J Pharmacol Exp Ther. 2023 Oct;387(1):100-110. doi: 10.1124/jpet.122.001538. Epub 2023 Jul 19. J Pharmacol Exp Ther. 2023. PMID: 37468286 Free PMC article. Review.
Cited by
-
An Imbalance in Histone Modifiers Induces tRNA-Cys-GCA Overexpression and tRF-27 Accumulation by Attenuating Promoter H3K27me3 in Primary Trastuzumab-Resistant Breast Cancer.Cancers (Basel). 2024 Mar 11;16(6):1118. doi: 10.3390/cancers16061118. Cancers (Basel). 2024. PMID: 38539453 Free PMC article.
References
-
- Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–50. - PubMed
-
- Leyland-Jones B. Trastuzumab: hopes and realities. The Lancet Oncology. 2002;3:137–44. - PubMed
-
- Valabrega G, Montemurro F, Aglietta M. Trastuzumab: mechanism of action, resistance and future perspectives in HER2-overexpressing breast cancer. Ann Oncol. 2007;18:977–84. - PubMed
-
- Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat Med. 2000;6:443–6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

