6-Gingerol modulates miRNAs and PODXL gene expression via methyltransferase enzymes in NB4 cells: an in silico and in vitro study
- PMID: 39112503
- PMCID: PMC11306743
- DOI: 10.1038/s41598-024-68069-4
6-Gingerol modulates miRNAs and PODXL gene expression via methyltransferase enzymes in NB4 cells: an in silico and in vitro study
Abstract
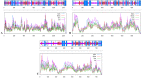
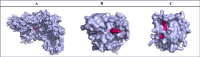
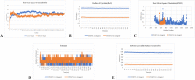
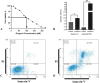
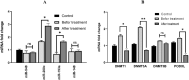
This investigation delves into the influence of predicted microRNAs on DNA methyltransferases (DNMTs) and the PODXL gene within the NB4 cell line, aiming to elucidate their roles in the pathogenesis of acute myeloid leukemia (AML). A comprehensive methodological framework was adopted to explore the therapeutic implications of 6-gingerol on DNMTs. This encompassed a suite of bioinformatics tools for protein structure prediction, docking, molecular dynamics, and ADMET profiling, alongside empirical assessments of miRNA and PODXL expression levels. Such a multifaceted strategy facilitated an in-depth understanding of 6-gingerol's potential efficacy in DNMT modulation. The findings indicate a nuanced interplay where 6-gingerol administration modulated miRNA expression levels, decreasing in DNMT1 and DNMT3A expression in NB4 cells. This alteration indirectly influenced PODXL expression, contributing to the manifestation of oncogenic phenotypes. The overexpression of DNMT1 and DNMT3A in NB4 cells may contribute to AML, which appears modulable via microRNAs such as miR-193a and miR-200c. Post-treatment with 6-gingerol, DNMT1 and DNMT3A expression alterations were observed, culminating in the upregulation of miR-193a and miR-200c. This cascade effect led to the dysregulation of tumor suppressor genes in cancer cells, including downregulation of PODXL, and the emergence of cancerous traits. These insights underscore the therapeutic promise of 6-gingerol in targeting DNMTs and microRNAs within the AML context.
Keywords: PODXL gene; AML; Docking; Methyltransferase; MiRNA; Molecular dynamic simulation; NB4 cell line.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Catechin-Induced changes in PODXL, DNMTs, and miRNA expression in Nalm6 cells: an integrated in silico and in vitro approach.BMC Complement Med Ther. 2024 Jun 15;24(1):234. doi: 10.1186/s12906-024-04521-2. BMC Complement Med Ther. 2024. PMID: 38879474 Free PMC article.
-
Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway.Blood. 2013 Jan 17;121(3):499-509. doi: 10.1182/blood-2012-07-444729. Epub 2012 Dec 6. Blood. 2013. PMID: 23223432
-
Overexpression of DNA (Cytosine-5)-Methyltransferase 1 (DNMT1) And DNA (Cytosine-5)-Methyltransferase 3A (DNMT3A) Is Associated with Aggressive Behavior and Hypermethylation of Tumor Suppressor Genes in Human Pituitary Adenomas.Med Sci Monit. 2018 Jul 13;24:4841-4850. doi: 10.12659/MSM.910608. Med Sci Monit. 2018. PMID: 30002361 Free PMC article.
-
DNA methyltransferases in hematological malignancies.J Genet Genomics. 2020 Jul 20;47(7):361-372. doi: 10.1016/j.jgg.2020.04.006. Epub 2020 Jul 24. J Genet Genomics. 2020. PMID: 32994141 Free PMC article. Review.
-
[Research progress on DNMT3A gene expression in Acute myeloid leukemia].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2024 Aug 10;41(8):1010-1015. doi: 10.3760/cma.j.cn511374-20210913-00746. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2024. PMID: 39097288 Review. Chinese.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

