P4-ATPase endosomal recycling relies on multiple retromer-dependent localization signals
- PMID: 39110530
- PMCID: PMC11481694
- DOI: 10.1091/mbc.E24-05-0209
P4-ATPase endosomal recycling relies on multiple retromer-dependent localization signals
Abstract
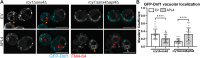
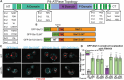
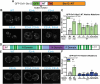
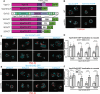
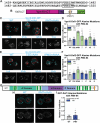
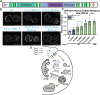
Type IV P-type ATPases (P4-ATPases) are lipid flippases that generate an asymmetric membrane organization essential for cell viability. The five budding yeast P4-ATPases traffic between the Golgi complex, plasma membrane, and endosomes but how they are recycled from the endolysosomal system to the Golgi complex is poorly understood. In this study, we find that P4-ATPase endosomal recycling is primarily driven by the retromer complex and the F-box protein Rcy1. Defects in P4-ATPase recycling result in their mislocalization to the vacuole and a substantial loss of membrane asymmetry. The P4-ATPases contain multiple predicted retromer sorting signals, and the characterization of these signals in Dnf1 and Dnf2 led to the identification of a novel retromer-dependent signal, IPM[ST] that acts redundantly with predicted motifs. Together, these results emphasize the importance of endosomal recycling for the functional localization of P4-ATPases and membrane organization.
Conflict of interest statement
Conflicts of interest: The authors declare no financial conflict of interest.
Figures




Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
"I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article.
-
Antioxidants for female subfertility.Cochrane Database Syst Rev. 2017 Jul 28;7(7):CD007807. doi: 10.1002/14651858.CD007807.pub3. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Aug 27;8:CD007807. doi: 10.1002/14651858.CD007807.pub4 PMID: 28752910 Free PMC article. Updated. Review.
References
-
- Aoki Y, Uenaka T, Aoki J, Umeda M, Inoue K (1994). A novel peptide probe for studying the transbilayer movement of phosphatidylethanolamine. J Biochem 116, 291–297. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

