Decoding human bone marrow hematopoietic stem and progenitor cells from fetal to birth
- PMID: 39108709
- PMCID: PMC11300922
- DOI: 10.1016/j.isci.2024.110445
Decoding human bone marrow hematopoietic stem and progenitor cells from fetal to birth
Abstract
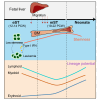
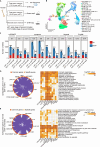
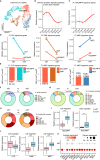
Bone marrow (BM) is the dominant site of hematopoiesis after 20 post-conception weeks (PCWs), but the intricacies of hematopoietic development in fetal BM up to birth and its involvement in malignancies remain unknown. Here, we compared the single-cell transcriptomic profile of BM hematopoietic stem and progenitor cells (HSPCs) at the early (12-14 PCW), middle (19-22 PCW) second trimester, and the neonatal stage. The stemness of hematopoietic stem cell and multipotent progenitor (HSC/MPP) is established at the middle second trimester, then maintained until birth. Furthermore, differentiation potentials toward three lineages are enhanced after the middle second trimester for birth, accompanied by the upregulation of aerobic metabolism. Notably, decreased stemness in HSCs/MPPs and higher interferon signals in progenitors at the early second trimester rendered the HSPCs more proximal to leukemogenesis. Collectively, our work elucidated the dynamics of fetal hematopoiesis in preparation for birth, offering valuable insights into the pathological processes underlying leukemia.
Keywords: Haematology; Molecular biology; Transcriptomics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Uncovering the emergence of HSCs in the human fetal bone marrow by single-cell RNA-seq analysis.Cell Stem Cell. 2022 Nov 3;29(11):1562-1579.e7. doi: 10.1016/j.stem.2022.10.005. Cell Stem Cell. 2022. PMID: 36332570
-
Murine fetal bone marrow does not support functional hematopoietic stem and progenitor cells until birth.Nat Commun. 2022 Sep 15;13(1):5403. doi: 10.1038/s41467-022-33092-4. Nat Commun. 2022. PMID: 36109585 Free PMC article.
-
Unique molecular and functional features of extramedullary hematopoietic stem and progenitor cell reservoirs in humans.Blood. 2022 Jun 9;139(23):3387-3401. doi: 10.1182/blood.2021013450. Blood. 2022. PMID: 35073399 Free PMC article.
-
Ionizing Particle Radiation as a Modulator of Endogenous Bone Marrow Cell Reprogramming: Implications for Hematological Cancers.Front Oncol. 2015 Oct 14;5:231. doi: 10.3389/fonc.2015.00231. eCollection 2015. Front Oncol. 2015. PMID: 26528440 Free PMC article. Review.
-
Regulation of hematopoietic and leukemic stem cells by the immune system.Cell Death Differ. 2015 Feb;22(2):187-98. doi: 10.1038/cdd.2014.89. Epub 2014 Jul 4. Cell Death Differ. 2015. PMID: 24992931 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

