The cap-binding complex modulates ABA-responsive transcript splicing during germination in barley (Hordeum vulgare)
- PMID: 39107424
- PMCID: PMC11303550
- DOI: 10.1038/s41598-024-69373-9
The cap-binding complex modulates ABA-responsive transcript splicing during germination in barley (Hordeum vulgare)
Abstract
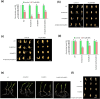
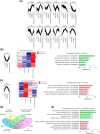
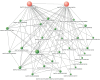
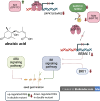
To decipher the molecular bases governing seed germination, this study presents the pivotal role of the cap-binding complex (CBC), comprising CBP20 and CBP80, in modulating the inhibitory effects of abscisic acid (ABA) in barley. Using both single and double barley mutants in genes encoding the CBC, we revealed that the double mutant hvcbp20.ab/hvcbp80.b displays ABA insensitivity, in stark contrast to the hypersensitivity observed in single mutants during germination. Our comprehensive transcriptome and metabolome analysis not only identified significant alterations in gene expression and splicing patterns but also underscored the regulatory nexus among CBC, ABA, and brassinosteroid (BR) signaling pathways.
Keywords: ABA; Alternative splicing; Barley; Cap-binding complex; Embryo; Germination; Transcriptome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Transcriptional regulatory programs underlying barley germination and regulatory functions of Gibberellin and abscisic acid.BMC Plant Biol. 2011 Jun 13;11:105. doi: 10.1186/1471-2229-11-105. BMC Plant Biol. 2011. PMID: 21668981 Free PMC article.
-
The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA.Plant Physiol. 2006 Jan;140(1):127-39. doi: 10.1104/pp.105.070318. Epub 2005 Dec 16. Plant Physiol. 2006. PMID: 16361522 Free PMC article.
-
Involvement of ABA in induction of secondary dormancy in barley (Hordeum vulgare L.) seeds.Plant Cell Physiol. 2008 Dec;49(12):1830-8. doi: 10.1093/pcp/pcn164. Epub 2008 Oct 30. Plant Cell Physiol. 2008. PMID: 18974197
-
mRNA cap binding proteins: effects on abscisic acid signal transduction, mRNA processing, and microarray analyses.Curr Top Microbiol Immunol. 2008;326:139-50. doi: 10.1007/978-3-540-76776-3_8. Curr Top Microbiol Immunol. 2008. PMID: 18630751 Review.
-
Alternative splicing in ABA signaling during seed germination.Front Plant Sci. 2023 Mar 16;14:1144990. doi: 10.3389/fpls.2023.1144990. eCollection 2023. Front Plant Sci. 2023. PMID: 37008485 Free PMC article. Review.
References
-
- Bradford, K. J. & Nonogaki, H. Seed Development, Dormancy and Germination (Blackwell Publishing Ltd, 2007). 10.1002/9780470988848.
-
- Boesewinkel, F. D. & Bouman, F. The seed: structure. In Embryology of Angiosperms (ed. Johri, B. M.) 567–610 (Springer Berlin Heidelberg, 1984). 10.1007/978-3-642-69302-1_12.
-
- Potokina, E., Sreenivasulu, N., Altschmied, L., Michalek, W. & Graner, A. Differential gene expression during seed germination in barley (Hordeumvulgare L.). Funct. Integr. Genom.2, 28–39 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

