ANGPTL4 Suppresses Clear Cell Renal Cell Carcinoma via Inhibition of Lysosomal Acid Lipase
- PMID: 39105498
- PMCID: PMC11348483
- DOI: 10.1158/2767-9764.CRC-24-0016
ANGPTL4 Suppresses Clear Cell Renal Cell Carcinoma via Inhibition of Lysosomal Acid Lipase
Abstract
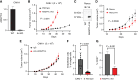
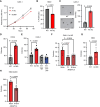
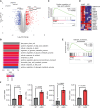
Renal cell carcinoma (RCC), the most common form of kidney cancer, is a heterogeneous disease with clear cell RCC (ccRCC) being the most prevalent and aggressive subtype. While most ccRCC tumors have elevated expression of angiopoietin-like4 (ANGPTL4), in our study we identified a significant subset of patients whose cancers show no increase in ANGPTL4 expression. These patients have a worse prognosis compared to the patients with high expression of ANGPTL4. These ANGPTL4-low cancers are characterized by the increased frequency of wild-type Von Hippel-Lindau(WT VHL), a gene that is commonly mutated in ccRCC, and an enrichment for genes associated with lipid metabolism. Using RCC tumor models with WT VHL, we demonstrate that ANGPTL4 behaves as a tumor suppressor. The loss of ANGPTL4 in ccRCC cell lines results in increased tumor growth and colony formation in a lysosomal acid lipase (LAL)-dependent manner, a phenotype rescued by the expression of N-terminus ANGPTL4. At the mechanistic level, the loss of ANGPTL4 increases LAL activity in ccRCC cells. These data suggest that ANGPTL4 enacts its tumor-suppressive effects in ccRCC by regulating LAL activity. Importantly, the identified patient cohort with low ANGPTL4 expression may exhibit increased reliance on lipid metabolism, which can be a point of target for future therapy.
Significance: Our data indicate angiopoietin-like 4 (ANGPTL4) acts as a tumor suppressor in clear cell renal cell carcinoma via regulating lipid metabolism and identifies a cohort of patients with lower expression of ANGPTL4 that are correlated with shorter survival.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
B.S. Davies reports grants from National Institutes of Health during the conduct of the study. W. Zhang reports a patent to PCT/US2020/057964 issued. R. Kolb reports grants from National Institute of Health, National Institute of Health, and National Institute of Health during the conduct of the study; in addition, R. Kolb has a patent number 62927289 issued. No disclosures were reported by the other authors.
Figures


Similar articles
-
Research on the effect of LAMP1 in the development and progression of ccRCC and its potential mechanism with LC3C-mediated autophagy.Front Immunol. 2024 Nov 28;15:1494005. doi: 10.3389/fimmu.2024.1494005. eCollection 2024. Front Immunol. 2024. PMID: 39669571 Free PMC article.
-
MYBL2 promotes proliferation of clear cell renal cell carcinoma by regulating TOP2A and activating AKT/mTOR signaling pathway.FASEB J. 2025 Jan 31;39(2):e70330. doi: 10.1096/fj.202401910R. FASEB J. 2025. PMID: 39831843
-
Mitochondrial respiratory complex II is altered in renal carcinoma.Biochim Biophys Acta Mol Basis Dis. 2025 Jan;1871(1):167556. doi: 10.1016/j.bbadis.2024.167556. Epub 2024 Oct 31. Biochim Biophys Acta Mol Basis Dis. 2025. PMID: 39486656
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Aggressiveness classification of clear cell renal cell carcinoma using registration-independent radiology-pathology correlation learning.Med Phys. 2025 Jan;52(1):300-320. doi: 10.1002/mp.17476. Epub 2024 Oct 24. Med Phys. 2025. PMID: 39447001 Free PMC article.
References
-
- Padala SA, Kallam A. Clear cell renal carcinoma. StatPearls. Treasure Island (FL): StatPearls Publishing; 2023. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

