Mismatch repair-proficient tumor footprints in the sands of immune desert: mechanistic constraints and precision platforms
- PMID: 39100682
- PMCID: PMC11294168
- DOI: 10.3389/fimmu.2024.1414376
Mismatch repair-proficient tumor footprints in the sands of immune desert: mechanistic constraints and precision platforms
Abstract
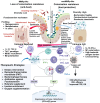
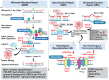
Mismatch repair proficient (MMRp) tumors of colorectal origin are one of the prevalent yet unpredictable clinical challenges. Despite earnest efforts, optimal treatment modalities have yet to emerge for this class. The poor prognosis and limited actionability of MMRp are ascribed to a low neoantigen burden and a desert-like microenvironment. This review focuses on the critical roadblocks orchestrated by an immune evasive mechanistic milieu in the context of MMRp. The low density of effector immune cells, their weak spatiotemporal underpinnings, and the high-handedness of the IL-17-TGF-β signaling are intertwined and present formidable challenges for the existing therapies. Microbiome niche decorated by Fusobacterium nucleatum alters the metabolic program to maintain an immunosuppressive state. We also highlight the evolving strategies to repolarize and reinvigorate this microenvironment. Reconstruction of anti-tumor chemokine signaling, rational drug combinations eliciting T cell activation, and reprograming the maladapted microbiome are exciting developments in this direction. Alternative vulnerability of other DNA damage repair pathways is gaining momentum. Integration of liquid biopsy and ex vivo functional platforms provide precision oncology insights. We illustrated the perspectives and changing landscape of MMRp-CRC. The emerging opportunities discussed in this review can turn the tide in favor of fighting the treatment dilemma for this elusive cancer.
Keywords: MMRp; colorectal cancer; functional platforms; gut microbiota; precision medicine; predictive biomarkers; therapeutic vulnerability; tumor immune microenvironment.
Copyright © 2024 Majumder, Nataraj, Maitreyi and Datta.
Conflict of interest statement
BM, LM and NN are employees of Bugworks Research Inc. and hold shares of the company. SD is co-founder of Bugworks Research Inc and holds equities. BM serves as an honorary scientific advisor of Praesidia Biotherapeutics, Bangalore, India and a consultant of Aryastha Life Sciences, Hyderabad, India.
Figures


Similar articles
-
Genomics and emerging biomarkers for immunotherapy of colorectal cancer.Semin Cancer Biol. 2018 Oct;52(Pt 2):189-197. doi: 10.1016/j.semcancer.2018.02.010. Epub 2018 Mar 1. Semin Cancer Biol. 2018. PMID: 29501787 Review.
-
Intratumoral Adaptive Immunosuppression and Type 17 Immunity in Mismatch Repair Proficient Colorectal Tumors.Clin Cancer Res. 2019 Sep 1;25(17):5250-5259. doi: 10.1158/1078-0432.CCR-19-0114. Epub 2019 May 6. Clin Cancer Res. 2019. PMID: 31061070 Free PMC article.
-
A phase 2 study of GVAX colon vaccine with cyclophosphamide and pembrolizumab in patients with mismatch repair proficient advanced colorectal cancer.Cancer Med. 2020 Feb;9(4):1485-1494. doi: 10.1002/cam4.2763. Epub 2019 Dec 26. Cancer Med. 2020. PMID: 31876399 Free PMC article. Clinical Trial.
-
Deficient Mismatch Repair and the Role of Immunotherapy in Metastatic Colorectal Cancer.Curr Treat Options Oncol. 2016 Aug;17(8):41. doi: 10.1007/s11864-016-0414-4. Curr Treat Options Oncol. 2016. PMID: 27315067 Review.
-
Neoantigen-specific immunity in low mutation burden colorectal cancers of the consensus molecular subtype 4.Genome Med. 2019 Dec 30;11(1):87. doi: 10.1186/s13073-019-0697-8. Genome Med. 2019. PMID: 31888734 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

