Emerging roles of ATG9/ATG9A in autophagy: implications for cell and neurobiology
- PMID: 39099167
- PMCID: PMC11572220
- DOI: 10.1080/15548627.2024.2384349
Emerging roles of ATG9/ATG9A in autophagy: implications for cell and neurobiology
Abstract
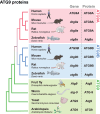
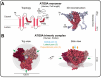
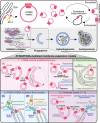
Atg9, the only transmembrane protein among many autophagy-related proteins, was first identified in the year 2000 in yeast. Two homologs of Atg9, ATG9A and ATG9B, have been found in mammals. While ATG9B shows a tissue-specific expression pattern, such as in the placenta and pituitary gland, ATG9A is ubiquitously expressed. Additionally, ATG9A deficiency leads to severe defects not only at the molecular and cellular levels but also at the organismal level, suggesting key and fundamental roles for ATG9A. The subcellular localization of ATG9A on small vesicles and its functional relevance to autophagy have suggested a potential role for ATG9A in the lipid supply during autophagosome biogenesis. Nevertheless, the precise role of ATG9A in the autophagic process has remained a long-standing mystery, especially in neurons. Recent findings, however, including structural, proteomic, and biochemical analyses, have provided new insights into its function in the expansion of the phagophore membrane. In this review, we aim to understand various aspects of ATG9 (in invertebrates and plants)/ATG9A (in mammals), including its localization, trafficking, and other functions, in nonneuronal cells and neurons by comparing recent discoveries related to ATG9/ATG9A and proposing directions for future research.Abbreviation: AP-4: adaptor protein complex 4; ATG: autophagy related; cKO: conditional knockout; CLA-1: CLArinet (functional homolog of cytomatrix at the active zone proteins piccolo and fife); cryo-EM: cryogenic electron microscopy; ER: endoplasmic reticulum; KO: knockout; PAS: phagophore assembly site; PtdIns3K: class III phosphatidylinositol 3-kinase; PtdIns3P: phosphatidylinositol-3-phosphate; RB1CC1/FIP200: RB1 inducible coiled-coil 1; SV: synaptic vesicle; TGN: trans-Golgi network; ULK: unc-51 like autophagy activating kinase; WIPI2: WD repeat domain, phosphoinositide interacting 2.
Keywords: ATG proteins; ATG9; ATG9A; autophagy; lipid scramblase; phagophore expansion.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
ZDHHC7-mediated S-palmitoylation of ATG16L1 facilitates LC3 lipidation and autophagosome formation.Autophagy. 2024 Dec;20(12):2719-2737. doi: 10.1080/15548627.2024.2386915. Epub 2024 Aug 11. Autophagy. 2024. PMID: 39087410
-
The differential expression patterns of Atg9a and Atg9b in cells of the reproductive organs.Clin Exp Reprod Med. 2024 Dec;51(4):301-308. doi: 10.5653/cerm.2023.06737. Epub 2024 May 17. Clin Exp Reprod Med. 2024. PMID: 38757275 Free PMC article.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
The Role of ATG9 Vesicles in Autophagosome Biogenesis.J Mol Biol. 2024 Aug 1;436(15):168489. doi: 10.1016/j.jmb.2024.168489. Epub 2024 Feb 10. J Mol Biol. 2024. PMID: 38342428 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
