Japanese encephalitis virus NS5 protein interacts with nucleolin to enhance the virus replication
- PMID: 39078257
- PMCID: PMC11334521
- DOI: 10.1128/jvi.00858-24
Japanese encephalitis virus NS5 protein interacts with nucleolin to enhance the virus replication
Abstract
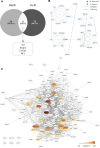
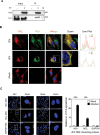
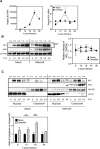
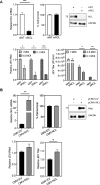
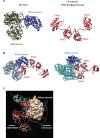
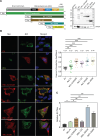
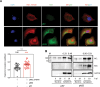
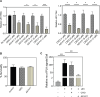
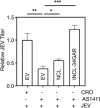
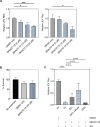
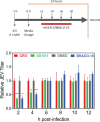
Japanese encephalitis virus (JEV) is an arthropod-borne, plus-strand flavivirus causing viral encephalitis in humans with a high case fatality rate. The JEV non-structural protein 5 (NS5) with the RNA-dependent RNA polymerase activity interacts with the viral and host proteins to constitute the replication complex. We have identified the multifunctional protein Nucleolin (NCL) as one of the several NS5-interacting host proteins. We demonstrate the interaction and colocalization of JEV NS5 with NCL in the virus-infected HeLa cells. The siRNA-mediated knockdown of NCL indicated that it was required for efficient viral replication. Importantly, JEV grew to higher titers in cells over-expressing exogenous NCL, demonstrating its pro-viral role. We demonstrated that NS5 interacted with the RRM and GAR domains of NCL. We show that the NCL-binding aptamer AS1411 containing the G-quadruplex (GQ) structure and the GQ ligand BRACO-19 caused significant inhibition of JEV replication. The antiviral effect of AS1411 and BRACO-19 could be overcome in HeLa cells by the overexpression of exogenous NCL. We demonstrated that the synthetic RNAs derived from the 3'-NCR of JEV genomic RNA containing the GQ sequence could bind NCL in vitro. The replication complex binding to the 3'-NCR is required for the viral RNA synthesis. It is likely that NCL present in the replication complex destabilizes the GQ structures in the genomic RNA, thus facilitating the movement of the replication complex resulting in efficient virus replication.IMPORTANCEJapanese encephalitis virus (JEV) is endemic in most parts of South-East Asia and the Western Pacific region, causing epidemics of encephalitis with a high case fatality rate. While a tissue culture-derived JEV vaccine is available, no antiviral therapy exists. The JEV NS5 protein has RNA-dependent RNA polymerase activity. Together with several host and viral proteins, it constitutes the replication complex necessary for virus replication. Understanding the interaction of NS5 with the host proteins could help design novel antivirals. We identified Nucleolin (NCL) as a crucial host protein interactor of JEV NS5 having a pro-viral role in virus replication. The NS5-interacting NCL binds to the G-quadruplex (GQ) structure sequence in the 3'-NCR of JEV RNA. This may smoothen the movement of the replication complex along the genomic RNA, thereby facilitating the virus replication. This study is the first report on how NCL, a host protein, helps in JEV replication through GQ-binding.
Keywords: RNA-dependent RNA polymerase; cytoplasmic translocation; flavivirus; nuclear protein; protein-protein interaction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Cellular NONO protein binds to the flavivirus replication complex and promotes positive-strand RNA synthesis.J Virol. 2024 Dec 17;98(12):e0029724. doi: 10.1128/jvi.00297-24. Epub 2024 Nov 5. J Virol. 2024. PMID: 39499073
-
Erratum: Eyestalk Ablation to Increase Ovarian Maturation in Mud Crabs.J Vis Exp. 2023 May 26;(195). doi: 10.3791/6561. J Vis Exp. 2023. PMID: 37235796
-
Alpha-synuclein expression in neurons modulates Japanese encephalitis virus infection.J Virol. 2024 Dec 17;98(12):e0041824. doi: 10.1128/jvi.00418-24. Epub 2024 Nov 7. J Virol. 2024. PMID: 39508602
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Exploring conceptual and theoretical frameworks for nurse practitioner education: a scoping review protocol.JBI Database System Rev Implement Rep. 2015 Oct;13(10):146-55. doi: 10.11124/jbisrir-2015-2150. JBI Database System Rev Implement Rep. 2015. PMID: 26571290
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

