Rabenosyn-5 suppresses non-small cell lung cancer metastasis via inhibiting CDC42 activity
- PMID: 39075137
- PMCID: PMC11489121
- DOI: 10.1038/s41417-024-00813-4
Rabenosyn-5 suppresses non-small cell lung cancer metastasis via inhibiting CDC42 activity
Abstract
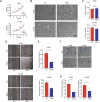
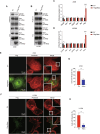
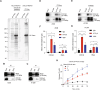
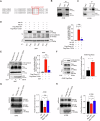
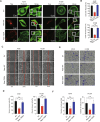
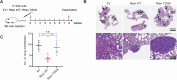
Metastasis, the primary cause of death in lung cancer patients, is facilitated by cytoskeleton remodeling, which plays a crucial role in cancer cell migration and invasion. However, the precise regulatory mechanisms of intracellular trafficking proteins involved in cytoskeleton remodeling remain unclear. In this study, we have identified Rabenosyn-5 (Rbsn) as an inhibitor of filopodia formation and lung cancer metastasis. Mechanistically, Rbsn interacts with CDC42 and functions as a GTPase activating protein (GAP), thereby inhibiting CDC42 activity and subsequent filopodia formation. Furthermore, we have discovered that Akt phosphorylates Rbsn at the Thr253 site, and this phosphorylation negates the inhibitory effect of Rbsn on CDC42 activity. Additionally, our analysis reveals that Rbsn expression is significantly downregulated in lung cancer, and this decrease is associated with a worse prognosis. These findings provide strong evidence supporting the role of Rbsn in suppressing lung cancer progression through the inhibition of metastasis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis.Oncotarget. 2015 Nov 3;6(34):36278-91. doi: 10.18632/oncotarget.5531. Oncotarget. 2015. PMID: 26462147 Free PMC article.
-
Expression analysis of Cdc42 in lung cancer and modulation of its expression by curcumin in lung cancer cell lines.Int J Oncol. 2012 May;40(5):1561-8. doi: 10.3892/ijo.2012.1336. Epub 2012 Jan 17. Int J Oncol. 2012. PMID: 22266952
-
Angio-associated migratory cell protein (AAMP) interacts with cell division cycle 42 (CDC42) and enhances migration and invasion in human non-small cell lung cancer cells.Cancer Lett. 2021 Apr 1;502:1-8. doi: 10.1016/j.canlet.2020.11.050. Epub 2020 Dec 3. Cancer Lett. 2021. PMID: 33279622
-
E-cadherin negatively regulates neoplastic growth in non-small cell lung cancer: role of Rho GTPases.Oncogene. 2010 May 13;29(19):2760-71. doi: 10.1038/onc.2010.39. Epub 2010 Mar 15. Oncogene. 2010. PMID: 20228844 Free PMC article.
-
miR-182 suppresses invadopodia formation and metastasis in non-small cell lung cancer by targeting cortactin gene.J Exp Clin Cancer Res. 2018 Jul 9;37(1):141. doi: 10.1186/s13046-018-0824-1. J Exp Clin Cancer Res. 2018. PMID: 29986736 Free PMC article.
References
-
- Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, et al. Non-small-cell lung cancer. Lancet. 2011;378:1727–40. - PubMed
-
- Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr, Wu YL, et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389:299–311. - PubMed
-
- Relli V, Trerotola M, Guerra E, Alberti S. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019;25:585–94. - PubMed
-
- Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv192–iv237. - PubMed
MeSH terms
Substances
Grants and funding
- LJKM20221787/Foundation of Liaoning Province Education Administration (Liaoning Province Education Administration Foundation)
- 2022JJ30364/Hunan Provincial Science and Technology Department (Department of Science and Technology of Hunan Province)
- 2020-MS-309/Natural Science Foundation of Liaoning Province (Liaoning Provincial Natural Science Foundation)
- 2022-NLTS-14-04/Natural Science Foundation of Liaoning Province (Liaoning Provincial Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

