Huntingtin contains an ubiquitin-binding domain and regulates lysosomal targeting of mitochondrial and RNA-binding proteins
- PMID: 39074279
- PMCID: PMC11317567
- DOI: 10.1073/pnas.2319091121
Huntingtin contains an ubiquitin-binding domain and regulates lysosomal targeting of mitochondrial and RNA-binding proteins
Abstract
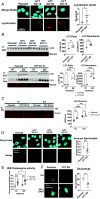
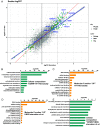
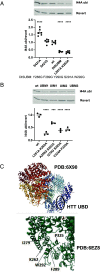
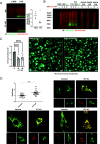
Understanding the normal function of the Huntingtin (HTT) protein is of significance in the design and implementation of therapeutic strategies for Huntington's disease (HD). Expansion of the CAG repeat in the HTT gene, encoding an expanded polyglutamine (polyQ) repeat within the HTT protein, causes HD and may compromise HTT's normal activity contributing to HD pathology. Here, we investigated the previously defined role of HTT in autophagy specifically through studying HTT's association with ubiquitin. We find that HTT interacts directly with ubiquitin in vitro. Tandem affinity purification was used to identify ubiquitinated and ubiquitin-associated proteins that copurify with a HTT N-terminal fragment under basal conditions. Copurification is enhanced by HTT polyQ expansion and reduced by mimicking HTT serine 421 phosphorylation. The identified HTT-interacting proteins include RNA-binding proteins (RBPs) involved in mRNA translation, proteins enriched in stress granules, the nuclear proteome, the defective ribosomal products (DRiPs) proteome and the brain-derived autophagosomal proteome. To determine whether the proteins interacting with HTT are autophagic targets, HTT knockout (KO) cells and immunoprecipitation of lysosomes were used to investigate autophagy in the absence of HTT. HTT KO was associated with reduced abundance of mitochondrial proteins in the lysosome, indicating a potential compromise in basal mitophagy, and increased lysosomal abundance of RBPs which may result from compensatory up-regulation of starvation-induced macroautophagy. We suggest HTT is critical for appropriate basal clearance of mitochondrial proteins and RBPs, hence reduced HTT proteostatic function with mutation may contribute to the neuropathology of HD.
Keywords: Huntingtin; RNA-binding proteins; autophagy; ubiquitin; ubiquitin-binding domain.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

Similar articles
-
Generation and Characterization of Knock-in Mouse Models Expressing Versions of Huntingtin with Either an N17 or a Combined PolyQ and Proline-Rich Region Deletion.J Huntingtons Dis. 2017;6(1):47-62. doi: 10.3233/JHD-160231. J Huntingtons Dis. 2017. PMID: 28211815 Free PMC article.
-
Mutant HTT (huntingtin) impairs mitophagy in a cellular model of Huntington disease.Autophagy. 2021 Mar;17(3):672-689. doi: 10.1080/15548627.2020.1728096. Epub 2020 Feb 24. Autophagy. 2021. PMID: 32093570 Free PMC article.
-
The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation.J Neurosci. 2014 Jan 22;34(4):1293-305. doi: 10.1523/JNEUROSCI.1870-13.2014. J Neurosci. 2014. PMID: 24453320 Free PMC article.
-
A Fresh Look at Huntingtin mRNA Processing in Huntington's Disease.J Huntingtons Dis. 2018;7(2):101-108. doi: 10.3233/JHD-180292. J Huntingtons Dis. 2018. PMID: 29865084 Free PMC article. Review.
-
Protective Proteolysis in Huntington's Disease: Unraveling the Role of Post-Translational Myristoylation of Huntingtin in Autophagy.J Huntingtons Dis. 2024;13(3):267-277. doi: 10.3233/JHD-240028. J Huntingtons Dis. 2024. PMID: 38995796 Free PMC article. Review.
Cited by
-
Methamphetamine Increases Tubulo-Vesicular Areas While Dissipating Proteins from Vesicles Involved in Cell Clearance.Int J Mol Sci. 2024 Sep 4;25(17):9601. doi: 10.3390/ijms25179601. Int J Mol Sci. 2024. PMID: 39273545 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- T32 GM008620/GM/NIGMS NIH HHS/United States
- NS116872/HHS | NIH | NIH Blueprint for Neuroscience Research (NIH Blueprint)
- R35GM148350/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- NS052789/HHS | NIH | NIH Blueprint for Neuroscience Research (NIH Blueprint)
- NSR37101996/HHS | NIH | NIH Blueprint for Neuroscience Research (NIH Blueprint)
- R35 NS116872/NS/NINDS NIH HHS/United States
- T32 AG000096/AG/NIA NIH HHS/United States
- R01 NS052789/NS/NINDS NIH HHS/United States
- F30 AG060704/AG/NIA NIH HHS/United States
- R35 GM148350/GM/NIGMS NIH HHS/United States
- T32GM008620/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- NS090390/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R37 NS101996/NS/NINDS NIH HHS/United States
- U54 NS091046/NS/NINDS NIH HHS/United States
- NS072453/HHS | NIH | NIH Blueprint for Neuroscience Research (NIH Blueprint)
- R01 NS090390/NS/NINDS NIH HHS/United States
- NS091046-01/HHS | NIH | NIH Blueprint for Neuroscience Research (NIH Blueprint)
- R56 NS090390/NS/NINDS NIH HHS/United States
- R01 NS072453/NS/NINDS NIH HHS/United States
- R35NS116872/HHS | NIH | NIH Blueprint for Neuroscience Research (NIH Blueprint)
- R35 GM145249/GM/NIGMS NIH HHS/United States
- T32AG000096/HHS | NIH | National Institute on Aging (NIA)
- R56 NS072453/NS/NINDS NIH HHS/United States
- R35GM145249/HHS | NIH | National Institute of General Medical Sciences (NIGMS)
- 1F30AG060704-01A1/HHS | NIH | National Institute on Aging (NIA)
LinkOut - more resources
Full Text Sources
Research Materials

