This is a preprint.
Experimental evolution of gene essentiality in bacteria
- PMID: 39071448
- PMCID: PMC11275930
- DOI: 10.1101/2024.07.16.600122
Experimental evolution of gene essentiality in bacteria
Abstract
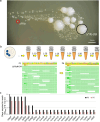
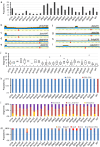
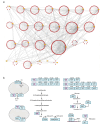
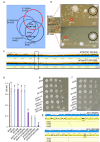
Essential gene products carry out fundamental cellular activities in interaction with other components. However, the lack of essential gene mutants and appropriate methodologies to link essential gene functions with their partners poses significant challenges. Here, we have generated deletion mutants in 32 genes previously identified as essential, with 23 mutants showing extremely slow growth in the SK36 strain of Streptococcus sanguinis. The 23 genes corresponding to these mutants encode components of diverse pathways, are widely conserved among bacteria, and are essential in many other bacterial species. Whole-genome sequencing of 243 independently evolved populations of these mutants has identified >1000 spontaneous suppressor mutations in experimental evolution. Many of these mutations define new gene and pathway relationships, such as F1Fo-ATPase/V1Vo-ATPase/TrkA1-H1 that were demonstrated across multiple Streptococcus species. Patterns of spontaneous mutations occurring in essential gene mutants differed from those found in wildtype. While gene duplications occurred rarely and appeared most often at later stages of evolution, substitutions, deletions, and insertions were prevalent in evolved populations. These essential gene deletion mutants and spontaneous mutations fixed in the mutant populations during evolution establish a foundation for understanding gene essentiality and the interaction of essential genes in networks.
Keywords: Experimental evolution; F1Fo-ATPase/V1Vo-ATPase/TrkA1-H1 gene pathway; Gene essentiality; Spontaneous mutations; Streptococcus; Whole-genome sequencing.
Conflict of interest statement
Declaration of interests A patent application has been submitted by P.X. and L.B. and the Virginia Commonwealth University based on these results.
Figures

Similar articles
-
Spontaneous Mutants of Streptococcus sanguinis with Defects in the Glucose-Phosphotransferase System Show Enhanced Post-Exponential-Phase Fitness.J Bacteriol. 2021 Oct 25;203(22):e0037521. doi: 10.1128/JB.00375-21. Epub 2021 Aug 30. J Bacteriol. 2021. PMID: 34460310 Free PMC article.
-
Identifying essential Streptococcus sanguinis genes using genome-wide deletion mutation.Methods Mol Biol. 2015;1279:15-23. doi: 10.1007/978-1-4939-2398-4_2. Methods Mol Biol. 2015. PMID: 25636610 Free PMC article.
-
Molecular and Functional Analysis of the Type IV Pilus Gene Cluster in Streptococcus sanguinis SK36.Appl Environ Microbiol. 2019 Mar 6;85(6):e02788-18. doi: 10.1128/AEM.02788-18. Print 2019 Mar 15. Appl Environ Microbiol. 2019. PMID: 30635384 Free PMC article.
-
Evolution of the Inhibitory and Non-Inhibitory ε, ζ, and IF1 Subunits of the F1FO-ATPase as Related to the Endosymbiotic Origin of Mitochondria.Microorganisms. 2022 Jul 7;10(7):1372. doi: 10.3390/microorganisms10071372. Microorganisms. 2022. PMID: 35889091 Free PMC article. Review.
-
Machine learning approach to gene essentiality prediction: a review.Brief Bioinform. 2021 Sep 2;22(5):bbab128. doi: 10.1093/bib/bbab128. Brief Bioinform. 2021. PMID: 33842944 Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
